Role of Structural and Dynamical Plasticity in Sin3: The Free PAH2 Domain is a Folded Module in mSin3B
van Ingen, H., Baltussen, M.A., Aelen, J., Vuister, G.W.(2006) J Mol Biology 358: 485-497
- PubMed: 16519900
- DOI: https://doi.org/10.1016/j.jmb.2006.01.100
- Primary Citation of Related Structures:
2F05 - PubMed Abstract:
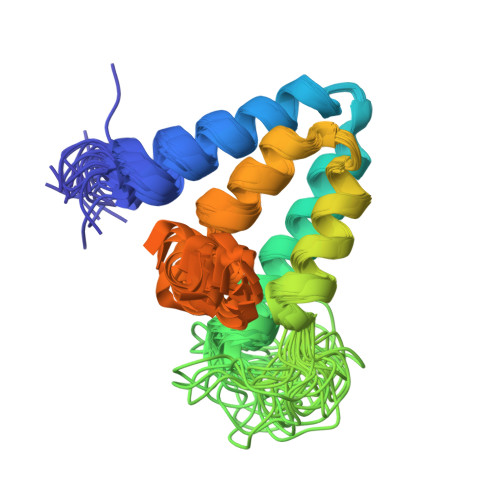
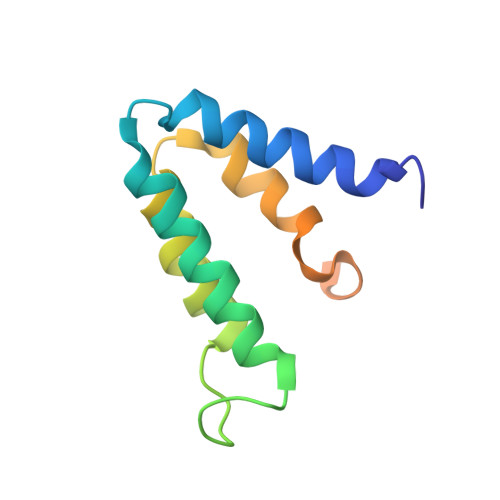
The co-repressor Sin3 is the essential scaffold protein of the Sin3/HDAC co-repressor complex, which is recruited to the DNA by a diverse group of transcriptional repressors, targeting genes involved in the regulation of the cell cycle, proliferation and differentiation. Sin3 contains four repeats commonly denoted as paired amphipathic helix (PAH1-4) domains that provide the principal interaction surface for various repressors. Here, we present the first structure of the free state of the PAH2 domain and discuss its implications for interaction with the repressors. The unbound conformation is very similar to the conformation observed when bound to either the Mad1 or HBP1 repressor, suggesting that the PAH2 domain serves as a template that guides proper folding of the unstructured repressor. The free PAH2 domain shows micro- to millisecond conformational exchange between the folded, major state and a partially unfolded, minor state. Upon complex formation, we observe a significant decrease in fast time-scale flexibility of local regions of the protein, correlated with the formation of intermolecular contacts, and an overall decrease in the slow time-scale conformational exchange. On the basis of our data and using a multiple sequence alignment of all PAH domains, we suggest that the PAH1, PAH2 and PAH3 domains form pre-folded binding modules in full-length Sin3 like beads-on-a-string, and act as folding templates for the interaction domains of their targets.
Organizational Affiliation:
Department of Physical Chemistry/Biophysical Chemistry, Institute for Molecules and Materials, Radboud University Nijmegen, Toernooiveld 1, 6525 ED Nijmegen, the Netherlands.