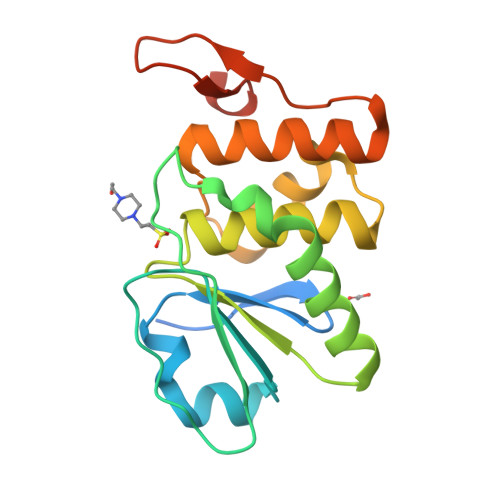
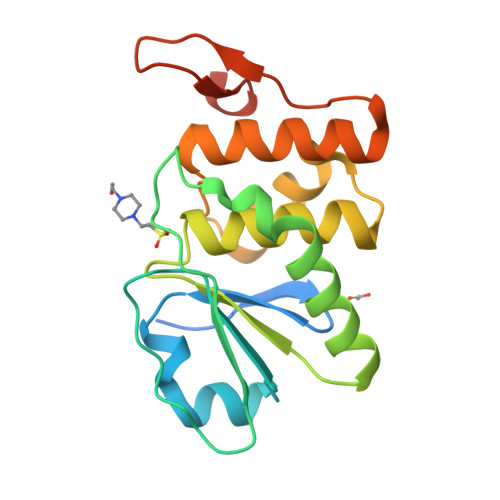
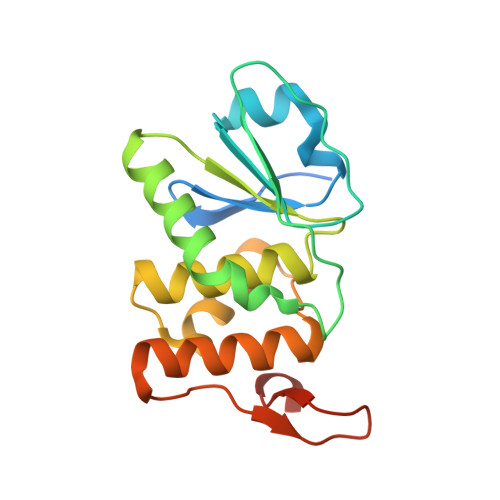
Structure of human DSP18, a member of the dual-specificity protein tyrosine phosphatase family.
Jeong, D.G., Cho, Y.H., Yoon, T.S., Kim, J.H., Son, J.H., Ryu, S.E., Kim, S.J.(2006) Acta Crystallogr D Biol Crystallogr 62: 582-588
- PubMed: 16699184
- DOI: https://doi.org/10.1107/S0907444906010109
- Primary Citation of Related Structures:
2ESB - PubMed Abstract:
The human dual-specificity protein phosphatase 18 (DSP18) gene and its protein product have recently been characterized. Like most DSPs, DSP18 displays dephosphorylating activity towards both phosphotyrosine and phosphothreonine residues. However, DSP18 is distinct from other known DSPs in terms of the existence of approximately 30 residues at the C-terminus of the catalytic domain and an unusual optimum activity profile at 328 K. The crystal structure of human DSP18 has been determined at 2.0 A resolution. The catalytic domain of DSP18 adopts a fold similar to that known for other DSP structures. Although good alignments are found with other DSPs, substantial differences are also found in the regions surrounding the active site, suggesting that DSP18 constitutes a unique structure with a distinct substrate specificity. Furthermore, the residues at the C-terminus fold into two antiparallel beta-strands and participate in extensive interactions with the catalytic domain, explaining the thermostability of DSP18.
Organizational Affiliation:
Systemic Proteomics Research Center, Korea Research Institute of Bioscience and Biotechnology, 52 Eoeun-Dong, Yuseong-Gu, Daejeon 305-333, South Korea.