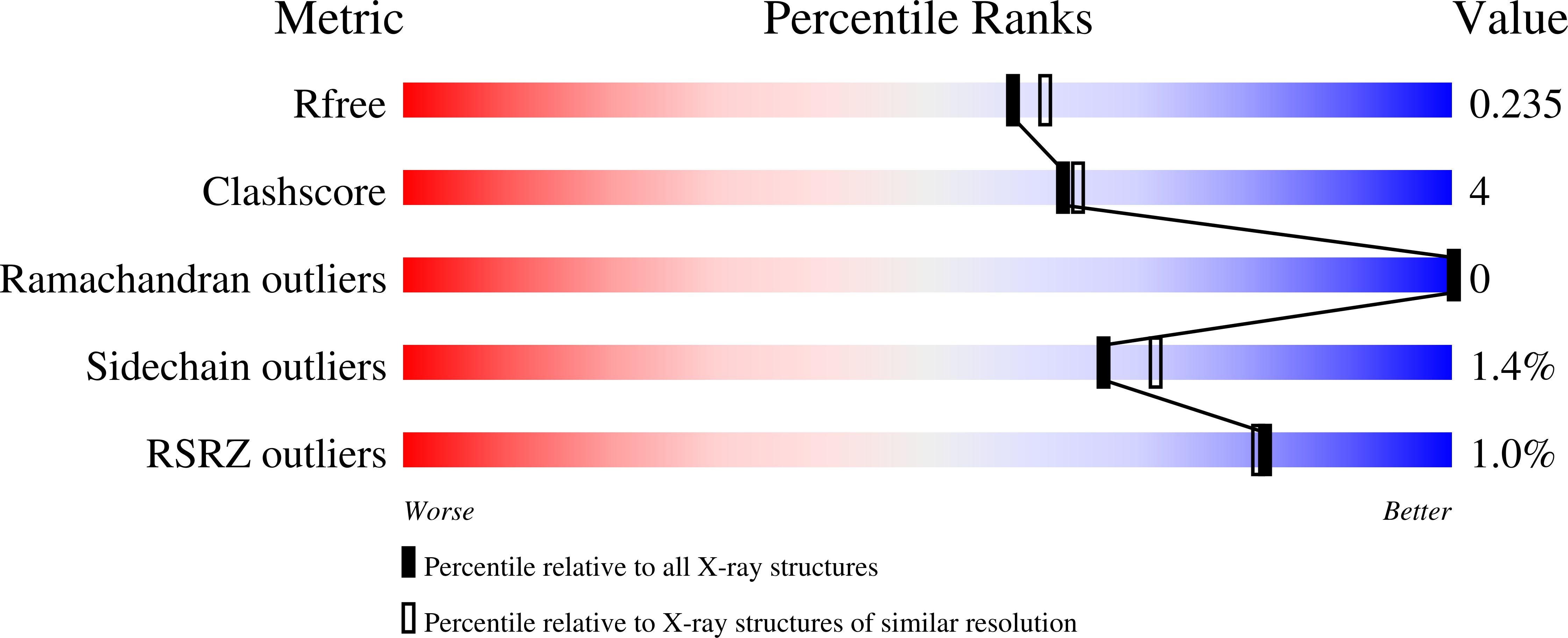
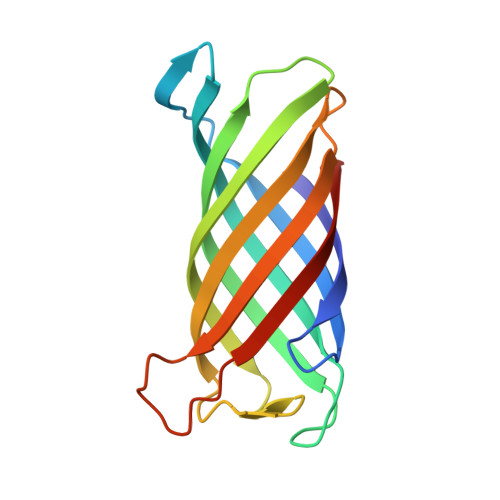
Crystal structure and catalytic mechanism of the LPS 3-O-deacylase PagL from Pseudomonas aeruginosa.
Rutten, L., Geurtsen, J., Lambert, W., Smolenaers, J.J., Bonvin, A.M., de Haan, A., van der Ley, P., Egmond, M.R., Gros, P., Tommassen, J.(2006) Proc Natl Acad Sci U S A 103: 7071-7076
- PubMed: 16632613
- DOI: https://doi.org/10.1073/pnas.0509392103
- Primary Citation of Related Structures:
2ERV - PubMed Abstract:
Pathogenic gram-negative bacteria can modify the lipid A portion of their lipopolysaccharide in response to environmental stimuli. 3-O-deacylation of lipid A by the outer membrane enzyme PagL modulates signaling through Toll-like receptor 4, leading to a reduced host immune response. We found that PagL is widely disseminated among gram-negative bacteria. Only four residues are conserved: a Ser, His, Phe, and Asn residue. Here, we describe the crystal structure of PagL from Pseudomonas aeruginosa to 2.0-A resolution. It consists of an eight-stranded beta-barrel with the axis tilted by approximately 30 degrees with respect to the lipid bilayer. The structure reveals that PagL contains an active site with a Ser-His-Glu catalytic triad and an oxyanion hole that comprises the conserved Asn. The importance of active site residues was confirmed in mutagenesis studies. Although PagL is most likely active as a monomer, its active site architecture shows high resemblance to that of the dimeric 12-stranded outer membrane phospholipase A. Modeling of the substrate lipid X onto the active site reveals that the 3-O-acyl chain is accommodated in a hydrophobic groove perpendicular to the membrane plane. In addition, an aspartate makes a hydrogen bond with the hydroxyl group of the 3-O-acyl chain, probably providing specificity of PagL toward lipid A.
Organizational Affiliation:
Department of Crystal and Structural Chemistry, Bijvoet Center for Biomolecular Research, Utrecht, The Netherlands.