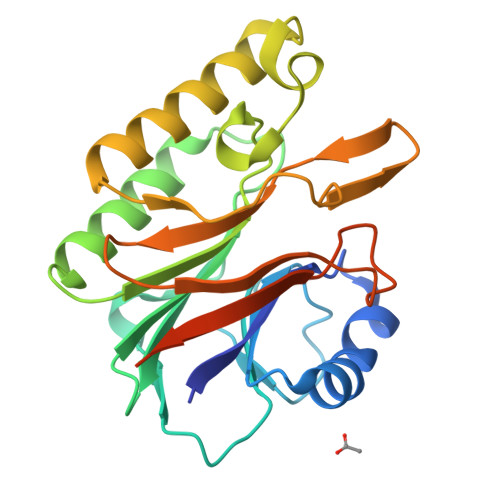
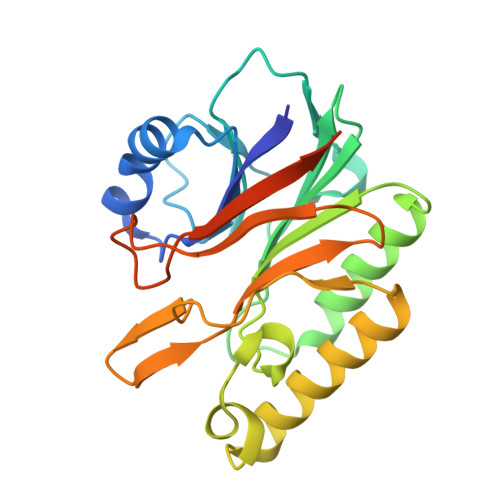
Characterization of the sequence specificity of the R1Bm endonuclease domain by structural and biochemical studies.
Maita, N., Aoyagi, H., Osanai, M., Shirakawa, M., Fujiwara, H.(2007) Nucleic Acids Res 35: 3918-3927
- PubMed: 17537809
- DOI: https://doi.org/10.1093/nar/gkm397
- Primary Citation of Related Structures:
2EI9 - PubMed Abstract:
R1Bm is a long interspersed element (LINE) inserted into a specific sequence within 28S rDNA of the silkworm genome. Of two open reading frames (ORFs) of R1Bm, ORF2 encodes a reverse transcriptase (RT) and an endonuclease (EN) domain which digests specifically both top and bottom strand of the target sequence in 28S rDNA. To elucidate the sequence specificity of EN domain of R1Bm (R1Bm EN), we examined the cleavage tendency for the target sequences, and found that 5'-A(G/C)(A/T)!(A/G)T-3' is the consensus sequence (! = cleavage site). We also determined the crystal structure of R1Bm EN at 2.0 A resolution. Its structure was basically similar to AP endonuclease family, but had a special beta-hairpin at the edge of the DNA binding surface, which is a common feature among EN of LINEs. Point-mutations on the DNA binding surface of R1Bm EN significantly decreased the cleavage activities, but did not affect the sequence recognition in most residues. However, two mutants Y98A and N180A had altered cleavage patterns, suggesting an important role of these residues (Y98 and N180) for the sequence recognition of R1Bm EN. In addition, Y98A mutant showed another cleavage pattern, that implies de novo design of novel sequence-specific EN.
Organizational Affiliation:
Graduate School of Systems Life Sciences, Kyushu University, Fukuoka 812-8582, Japan.