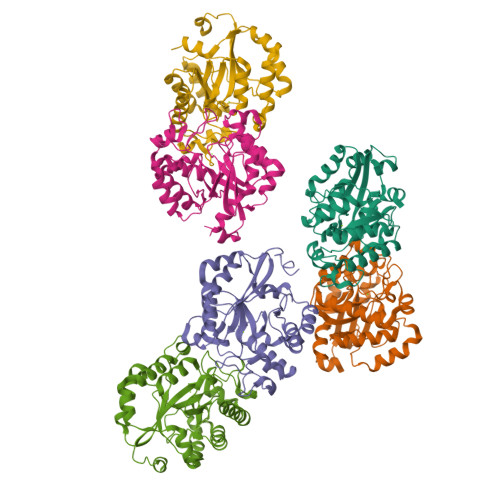
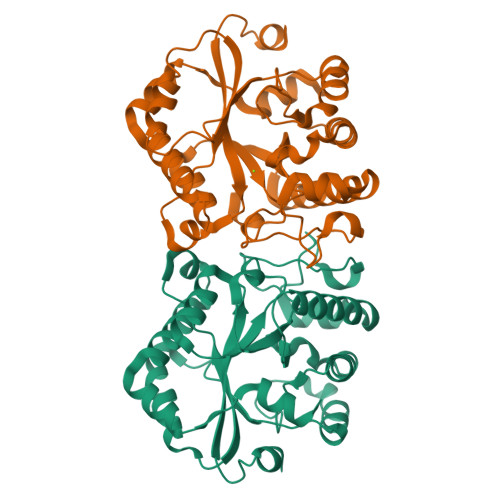
Crystal structure of the YdjC-family protein TTHB029 from Thermus thermophilus HB8: structural relationship with peptidoglycan N-acetylglucosamine deacetylase.
Imagawa, T., Iino, H., Kanagawa, M., Ebihara, A., Kuramitsu, S., Tsuge, H.(2008) Biochem Biophys Res Commun 367: 535-541
- PubMed: 18177738
- DOI: https://doi.org/10.1016/j.bbrc.2007.12.144
- Primary Citation of Related Structures:
2E67 - PubMed Abstract:
The YdjC-family protein is widely distributed, from human to bacteria, but so far no three-dimensional structure and functional analysis of this family of proteins has been reported. We determined the three-dimensional structure of the YdjC homolog TTHB029 at a resolution of 2.9A. The overall structure of the monomer consists of (betaalpha)-barrel fold forming a homodimer. Asp21, His60, and His127 residues coordinate to Mg(2+) as a possible active site. TTHB029 shows structural similarity to the peptidoglycan N-acetylglucosamine deacetylase from Streptococcus pneumoniae (SpPgdA). The active site groove of SpPgdA includes the Zn(2+) coordinated to Asp276, His326, and His330. Despite the low sequence identity, metal-binding residues of Asp-His-His were conserved among the two enzymes. There were definitive differences, however, in that one of the histidines of the metal-binding site was substituted for the other histidine located on the other loop. Moreover, these important metal-binding residues and the residues of the presumed active site are fully conserved in YdjC-family protein.
Organizational Affiliation:
Institute for Health Sciences, Tokushima Bunri University, 180 Nishihama, Yamashiro, Tokushima 770-8514, Japan.