Crystal Structure of the Vitamin B(12) Biosynthetic Cobaltochelatase, CbiX (S), from Archaeoglobus Fulgidus
Yin, J., Xu, L.X., Cherney, M.M., Raux-Deery, E., Bindley, A.A., Savchenko, A., Walker, J.R., Cuff, M.E., Warren, M.J., James, M.N.G.(2006) J Struct Funct Genomics 7: 37-50
- PubMed: 16835730
- DOI: https://doi.org/10.1007/s10969-006-9008-x
- Primary Citation of Related Structures:
2DJ5 - PubMed Abstract:
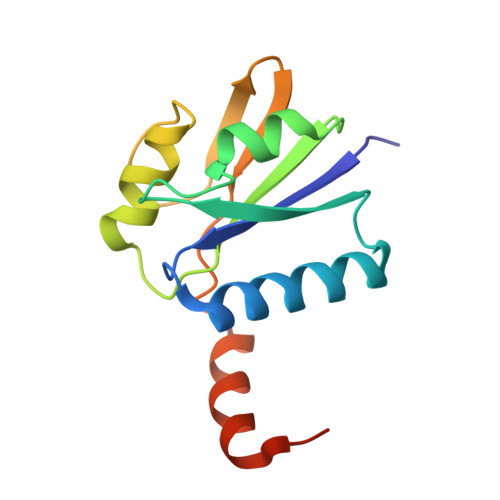
The Archaeoglobus fulgidus gene af0721 encodes CbiX(S), a small cobaltochelatase associated with the anaerobic biosynthesis of vitamin B12 (cobalamin). The protein was shown to have activity both in vivo and in vitro, catalyzing the insertion of Co2+ into sirohydrochlorin. The structure of CbiX(S) was determined in two different crystal forms and was shown to consist of a central mixed beta-sheet flanked by four alpha-helices, one of which originates in the C-terminus of a neighboring molecule. CbiX(S) is about half the size of other Class II tetrapyrrole chelatases. The overall topography of CbiX(S) exhibits substantial resemblance to both the N- and C-terminal regions of several members of the Class II metal chelatases involved in tetrapyrrole biosynthesis. Two histidines (His10 and His74), are in similar positions as the catalytic histidine residues in the anaerobic cobaltochelatase CbiK (His145 and His207). In light of the hypothesis that suggests the larger chelatases evolved via gene duplication and fusion from a CbiX(S)-like enzyme, the structure of AF0721 may represent that of an "ancestral" precursor of class II metal chelatases.
Organizational Affiliation:
Group in Protein Structure and Function, Department of Biochemistry, University of Alberta, Edmonton, AB, Canada, T6G 2H7.