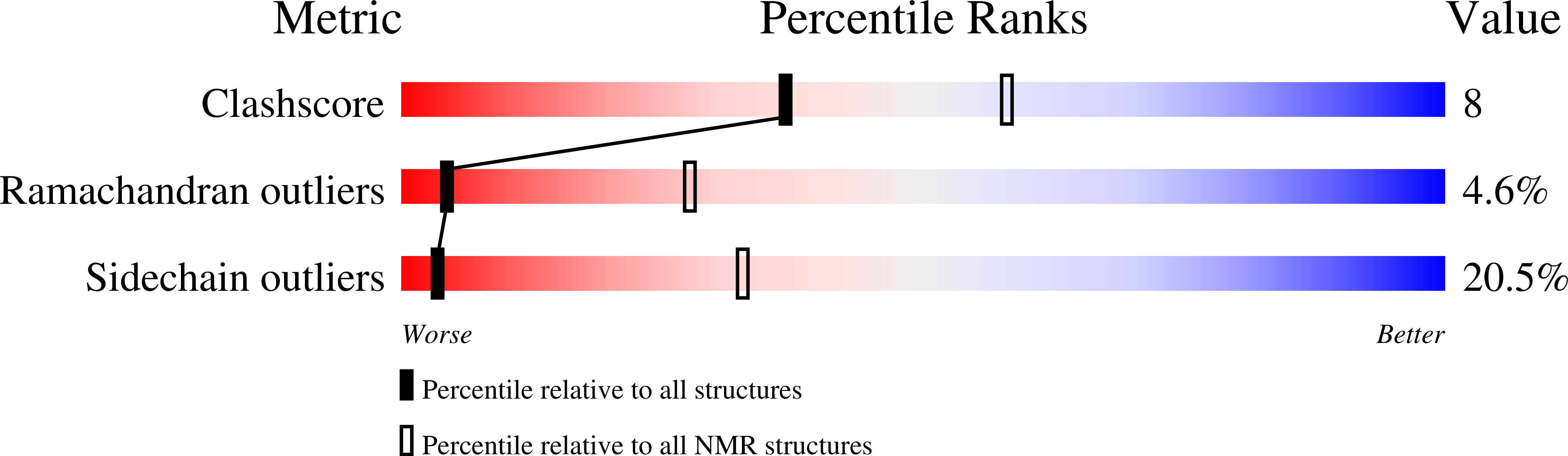The solution structure of horseshoe crab antimicrobial peptide tachystatin B with an inhibitory cystine-knot motif
Fujitani, N., Kouno, T., Nakahara, T., Takaya, K., Osaki, T., Kawabata, S., Mizuguchi, M., Aizawa, T., Demura, M., Nishimura, S., Kawano, K.(2007) J Pept Sci 13: 269-279
- PubMed: 17394123
- DOI: https://doi.org/10.1002/psc.846
- Primary Citation of Related Structures:
2DCV, 2DCW - PubMed Abstract:
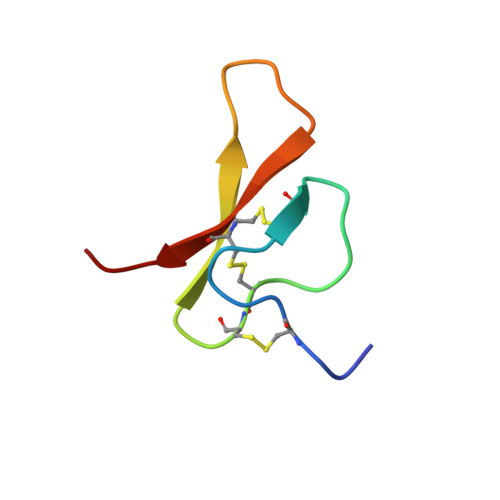
Tachystatin B is an antimicrobial and a chitin-binding peptide isolated from the Japanese horseshoe crab (Tachypleus tridentatus) consisting of two isopeptides called tachystatin B1 and B2. We have determined their solution structures using NMR experiments and distance geometry calculations. The 20 best converged structures of tachystatin B1 and B2 exhibited root mean square deviations of 0.46 and 0.49 A, respectively, for the backbone atoms in Cys(4)-Arg(40). Both structures have identical conformations, and they contain a short antiparallel beta-sheet with an inhibitory cystine-knot (ICK) motif that is distributed widely in the antagonists for voltage-gated ion channels, although tachystatin B does not have neurotoxic activity. The structural homology search provided several peptides with structures similar to that of tachystatin B. However, most of them have the advanced functions such as insecticidal activity, suggesting that tachystatin B may be a kind of ancestor of antimicrobial peptide in the molecular evolutionary history. Tachystatin B also displays a significant structural similarity to tachystatin A, which is member of the tachystatin family. The structural comparison of both tachystatins indicated that Tyr(14) and Arg(17) in the long loop between the first and second strands might be the essential residues for binding to chitin.
Organizational Affiliation:
Division of Advanced Chemical Biology, Graduate School of Advanced Life Science, Frontier Research Center for Post-Genome Science and Technology, Hokkaido University, Sapporo 001-0021, Japan.