Conformation of Amyloid Fibrils of beta2-Microglobulin Probed by Tryptophan Mutagenesis
Kihara, M., Chatani, E., Iwata, K., Yamamoto, K., Matsuura, T., Nakagawa, A., Naiki, H., Goto, Y.(2006) J Biological Chem 281: 31061-31069
- PubMed: 16901902
- DOI: https://doi.org/10.1074/jbc.M605358200
- Primary Citation of Related Structures:
2D4D, 2D4F - PubMed Abstract:
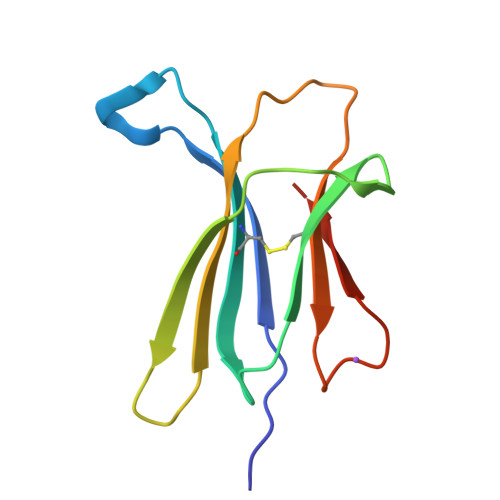
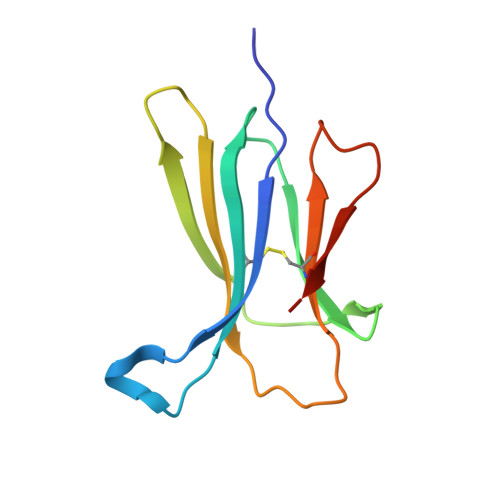
Beta2-microglobulin (beta2-m), a protein responsible for dialysis-related amyloidosis, adopts an immunoglobulin domain fold in its native state. Although beta2-m has Trp residues at positions 60 and 95, both are located near the surface of the domain. Hence, beta2-m does not have a conserved Trp common to other immunoglobulin domains, which is buried in close proximity to the disulfide bond. To study the structure of amyloid fibrils in relation to their native fold, we prepared a series of Trp mutants. Trp60 and Trp95 were both replaced with Phe, and a single Trp was introduced at various positions. Among various mutants, W39-beta2-m, in which a Trp was introduced at the position corresponding to the conserved Trp, exhibited a remarkable quenching of fluorescence in the native state, as observed for other immunoglobulin domains. An x-ray structural analysis revealed that W39-beta2-m assumes the native fold with Trp39 located in the vicinity of the disulfide bond. Comparison of the fluorescence spectra of various mutants for the native and fibrillar forms indicated that, while the Trp residues introduced in the middle of the beta2-m sequence tend to be buried in the fibrils, those located in the C-terminal region are more exposed. In addition, the fluorescence spectra of fibrils prepared at pH 2.5 and 7.0 revealed a large difference in the fluorescence intensity for W60-beta2-m, implying a major structural difference between them.
Organizational Affiliation:
Institute for Protein Research, Osaka University, and CREST, Japan Science and Technology Agency, Yamadaoka 3-2, Suita, Osaka 565-0871, Japan.