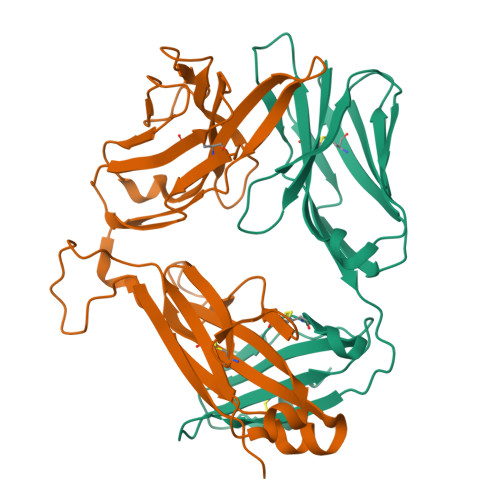
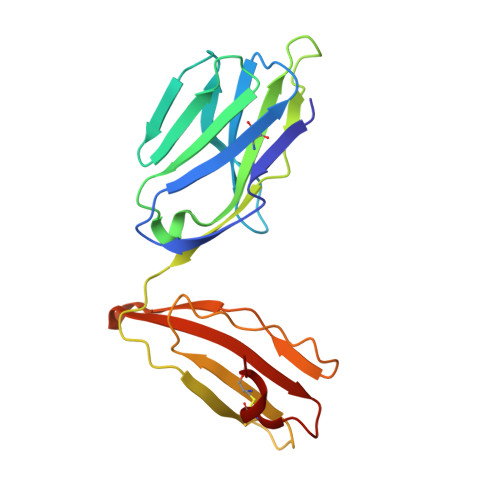
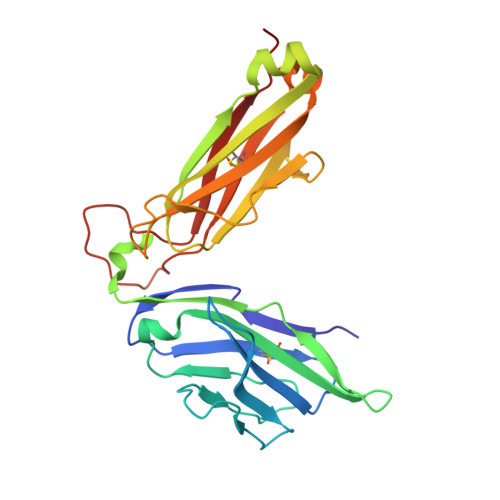
Structrue and Binding Kinetics of Three Different Human Cd1D-Alpha-Galactosylceramide-Specific T Cell Receptors
Gadola, S.D., Koch, M., Marles-Wright, J., Lissin, N.M., Sheperd, D., Matulis, G., Harlos, K., Villiger, P.M., Stuart, D.I., Jakobsen, B.K., Cerundolo, V., Jones, E.Y.(2006) J Exp Medicine 203: 699
- PubMed: 16520393
- DOI: https://doi.org/10.1084/jem.20052369
- Primary Citation of Related Structures:
2CDE, 2CDF, 2CDG - PubMed Abstract:
Invariant human TCR Valpha24-Jalpha18+/Vbeta11+ NKT cells (iNKT) are restricted by CD1d-alpha-glycosylceramides. We analyzed crystal structures and binding characteristics for an iNKT TCR plus two CD1d-alpha-GalCer-specific Vbeta11+ TCRs that use different TCR Valpha chains. The results were similar to those previously reported for MHC-peptide-specific TCRs, illustrating the versatility of the TCR platform. Docking TCR and CD1d-alpha-GalCer structures provided plausible insights into their interaction. The model supports a diagonal orientation of TCR on CD1d and suggests that complementarity determining region (CDR)3alpha, CDR3beta, and CDR1beta interact with ligands presented by CD1d, whereas CDR2beta binds to the CD1d alpha1 helix. This docking provides an explanation for the dominant usage of Vbeta11 and Vbeta8.2 chains by human and mouse iNKT cells, respectively, for recognition of CD1d-alpha-GalCer.
Organizational Affiliation:
Department of Rheumathology and Clinical Immunology, University of Bern, Inselspital, Berne CH-3010, Switzerland. stephan.gadola@insel.ch