Structure of the Functional Form of the Mosquito Larvicidal Cry4Aa Toxin from Bacillus Thuringiensis at 2.8-Angstrom Resolution.
Boonserm, P., Mo, M., Angsuthanasombat, C., Lescar, J.(2006) J Bacteriol 188: 3391
- PubMed: 16621834
- DOI: https://doi.org/10.1128/JB.188.9.3391-3401.2006
- Primary Citation of Related Structures:
2C9K - PubMed Abstract:
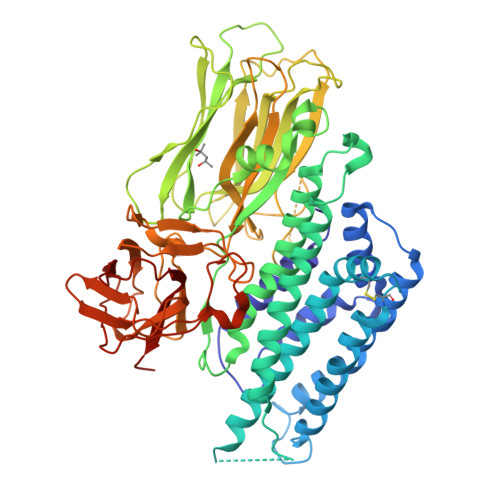
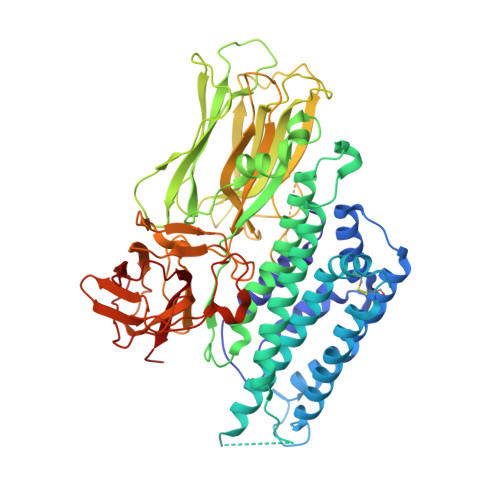
The Cry4Aa delta-endotoxin from Bacillus thuringiensis is toxic to larvae of Culex, Anopheles, and Aedes mosquitoes, which are vectors of important human tropical diseases. With the objective of designing modified toxins with improved potency that could be used as biopesticides, we determined the structure of this toxin in its functional form at a resolution of 2.8 angstroms. Like other Cry delta-endotoxins, the activated Cry4Aa toxin consists of three globular domains, a seven-alpha-helix bundle responsible for pore formation (domain I) and the following two other domains having structural similarities with carbohydrate binding proteins: a beta-prism (domain II) and a plant lectin-like beta-sandwich (domain III). We also studied the effect on toxicity of amino acid substitutions and deletions in three loops located at the surface of the putative receptor binding domain II of Cry4Aa. Our results indicate that one loop is an important determinant of toxicity, presumably through attachment of Cry4Aa to the surface of mosquito cells. The availability of the Cry4Aa structure should guide further investigations aimed at the molecular basis of the target specificity and membrane insertion of Cry endotoxins.
Organizational Affiliation:
Institute of Molecular Biology and Genetics, Mahidol University, Salaya Campus, Nakornpathom 73170, Thailand. mbpbs@mahidol.ac.th