Molecular Architecture and Ligand Recognition Determinants for T4 RNA Ligase
El Omari, K., Ren, J., Bird, L.E., Bona, M.K., Klarmann, G., Legrice, S.F.J., Stammers, D.K.(2006) J Biological Chem 281: 1573
- PubMed: 16263720
- DOI: https://doi.org/10.1074/jbc.M509658200
- Primary Citation of Related Structures:
2C5U - PubMed Abstract:
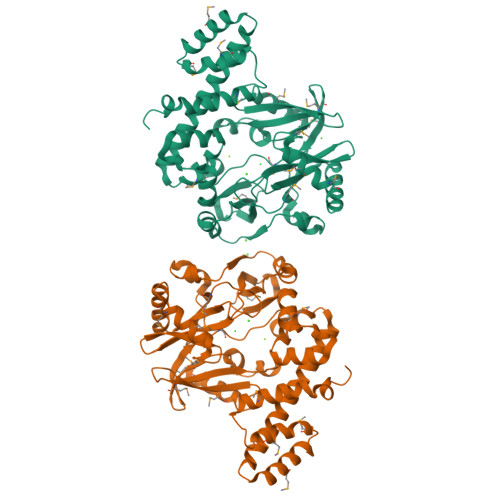
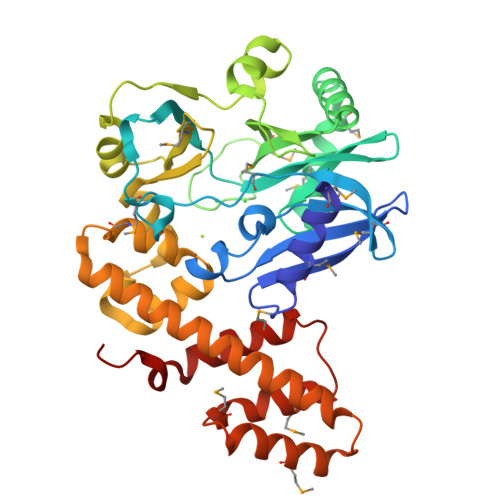
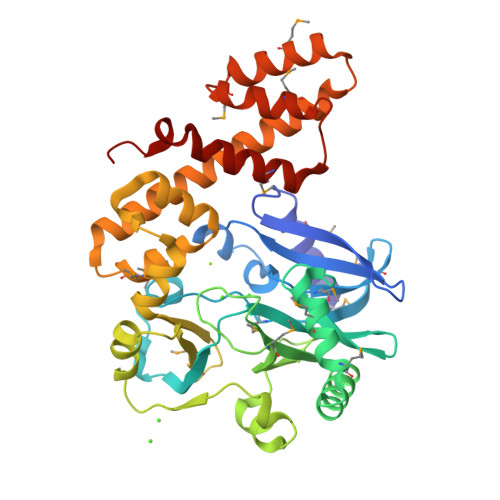
RNA ligase type 1 from bacteriophage T4 (Rnl1) is involved in countering a host defense mechanism by repairing 5'-PO4 and 3'-OH groups in tRNA(Lys). Rnl1 is widely used as a reagent in molecular biology. Although many structures for DNA ligases are available, only fragments of RNA ligases such as Rnl2 are known. We report the first crystal structure of a complete RNA ligase, Rnl1, in complex with adenosine 5'-(alpha,beta-methylenetriphosphate) (AMPcPP). The N-terminal domain is related to the equivalent region of DNA ligases and Rnl2 and binds AMPcPP but with further interactions from the additional N-terminal 70 amino acids in Rnl1 (via Tyr37 and Arg54) and the C-terminal domain (Gly269 and Asp272). The active site contains two metal ions, consistent with the two-magnesium ion catalytic mechanism. The C-terminal domain represents a new all alpha-helical fold and has a charge distribution and architecture for helix-nucleic acid groove interaction compatible with tRNA binding.
Organizational Affiliation:
Division of Structural Biology, The Wellcome Trust Centre for Human Genetics, University of Oxford, Roosevelt Drive, Oxford, OX3 7BN, UK.