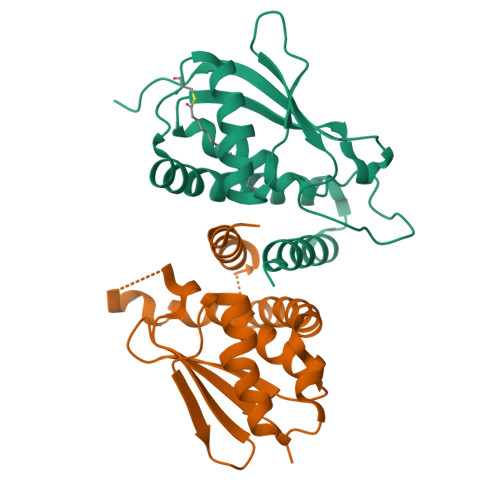
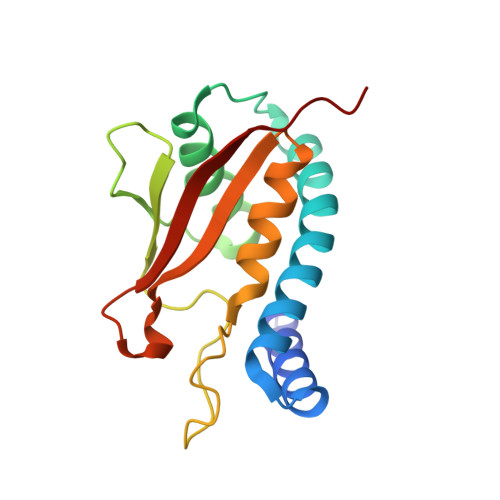
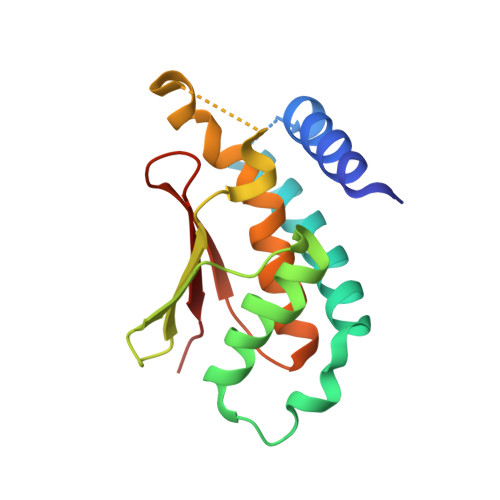
Biochemical and Crystallographic Studies Reveal a Specific Interaction between Trapp Subunits Trs33P and Bet3P
Kim, M.-S., Yi, M.-J., Lee, K.-H., Wagner, J., Munger, C., Kim, Y.-G., Whiteway, M., Cygler, M., Oh, B.-H., Sacher, M.(2005) Traffic 6: 1183
- PubMed: 16262728
- DOI: https://doi.org/10.1111/j.1600-0854.2005.00352.x
- Primary Citation of Related Structures:
2C0J - PubMed Abstract:
Transport protein particle (TRAPP) comprises a family of two highly related multiprotein complexes, with seven common subunits, that serve to target different classes of transport vesicles to their appropriate compartments. Defining the architecture of the complexes will advance our understanding of the functional differences between these highly related molecular machines. Genetic analyses in yeast suggested a specific interaction between the TRAPP subunits Bet3p and Trs33p. A mammalian bet3-trs33 complex was crystallized, and the structure was solved to 2.2 angstroms resolution. Intriguingly, the overall fold of the bet3 and trs33 monomers was similar, although the proteins had little overall sequence identity. In vitro experiments using yeast TRAPP subunits indicated that Bet3p binding to Trs33p facilitates the interaction between Bet3p and another TRAPP subunit, Bet5p. Mutational analysis suggests that yeast Trs33p facilitates other Bet3p protein-protein interactions. Furthermore, we show that Trs33p can increase the Golgi-localized pool of a mutated Bet3 protein normally found in the cytosol. We propose that one of the roles of Trs33p is to facilitate the incorporation of the Bet3p subunit into assembling TRAPP complexes.
Organizational Affiliation:
Center for Biomolecular Recognition, Department of Life Sciences and Division of Molecular and Life Sciences, Pohang University of Science and Technology, Pohang, Kyungbuk, 790-784, Korea.