3-(5-Chloro-2,4-Dihydroxyphenyl)-Pyrazole-4-Carboxamides as Inhibitors of the Hsp90 Molecular Chaperone.
Brough, P.A., Barril, X., Beswick, M., Dymock, B.W., Drysdale, M.J., Wright, L., Grant, K., Massey, A., Surgenor, A., Workman, P.(2005) Bioorg Med Chem Lett 15: 5197
- PubMed: 16213716
- DOI: https://doi.org/10.1016/j.bmcl.2005.08.091
- Primary Citation of Related Structures:
2BYH, 2BYI - PubMed Abstract:
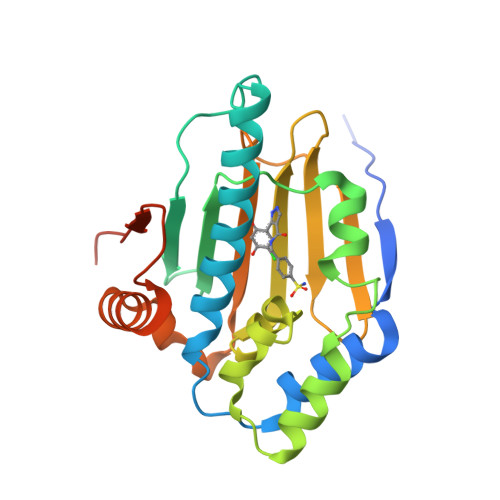
Information from X-ray crystal structures of Hsp90 inhibitors bound to the human Hsp90 molecular chaperone was used to assist in the design of 3-(5-chloro-2,4-dihydroxyphenyl)-pyrazole-4-carboxamides as novel inhibitors of Hsp90. Accessing an extra interaction with the protein via Phe138 gave a significant increase in binding potency compared to similar analogues that do not make this interaction.
Organizational Affiliation:
Vernalis Ltd, Granta Park, Great Abington, Cambridge CB1 6GB, UK. p.brough@vernalis.com