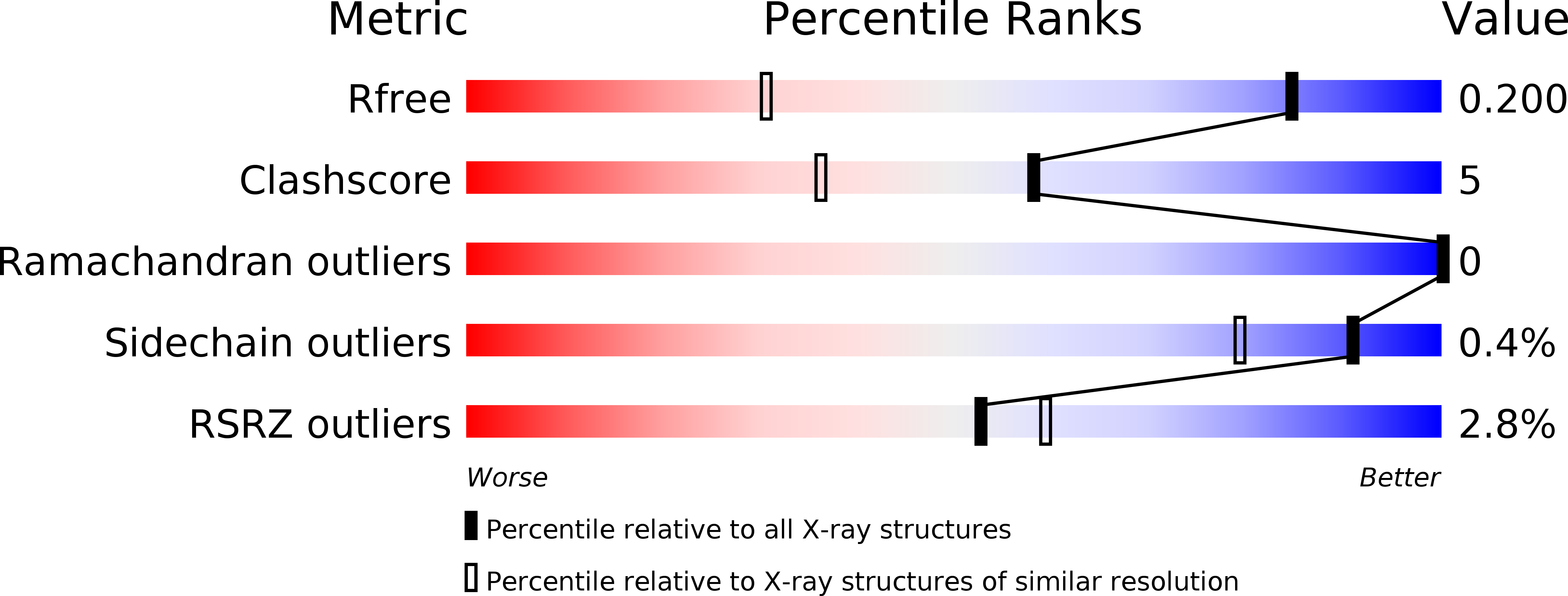
Versatile Peroxidase Oxidation of High Redox Potential Aromatic Compounds: Site-Directed Mutagenesis, Spectroscopic and Crystallographic Investigation of Three Long-Range Electron Transfer Pathways.
Perez-Boada, M., Ruiz-Duenas, F.J., Pogni, R., Basosi, R., Choinowski, T., Martinez, M.J., Piontek, K., Martinez, A.T.(2005) J Mol Biol 354: 385
- PubMed: 16246366
- DOI: https://doi.org/10.1016/j.jmb.2005.09.047
- Primary Citation of Related Structures:
2BOQ - PubMed Abstract:
Versatile peroxidases (VP), a recently described family of ligninolytic peroxidases, show a hybrid molecular architecture combining different oxidation sites connected to the heme cofactor. High-resolution crystal structures as well as homology models of VP isoenzymes from the fungus Pleurotus eryngii revealed three possibilities for long-range electron transfer for the oxidation of high redox potential aromatic compounds. The possible pathways would start either at Trp164 or His232 of isoenzyme VPL, and at His82 or Trp170 of isoenzyme VPS1. These residues are exposed, and less than 11 A apart from the heme. With the purpose of investigating their functionality, two single mutations (W164S and H232F) and one double mutation (W164S/P76H) were introduced in VPL that: (i) removed the two pathways in this isoenzyme; and (ii) incorporated the absent putative pathway. Analysis of the variants showed that Trp164 is required for oxidation of two high redox potential model substrates (veratryl alcohol and Reactive Black 5), whereas the two other pathways (starting at His232 and His82) are not involved in long-range electron transfer (LRET). None of the mutations affected Mn2+ oxidation, which would take place at the opposite side of the enzyme. Substitution of Trp164 by His also resulted in an inactive variant, indicating that an indole side-chain is required for activity. It is proposed that substrate oxidation occurs via a protein-based radical. For the first time in a ligninolytic peroxidase such an intermediate species could be detected by low-temperature electron paramagnetic resonance of H2O2-activated VP, and was found to exist at Trp164 as a neutral radical. The H2O2-activated VP was self-reduced in the absence of reducing substrates. Trp164 is also involved in this reaction, which in the W164S variant was blocked at the level of compound II. When analyzing VP crystal structures close to atomic resolution, no hydroxylation of the Trp164 Cbeta atom was observed (even after addition of several equivalents of H2O2). This is in contrast to lignin peroxidase Trp171. Analysis of the crystal structures of both peroxidases showed differences in the environment of the protein radical-forming residue that could affect its reactivity. These variations would also explain differences found for the oxidation of some high redox potential aromatic substrates.
Organizational Affiliation:
Centro de Investigaciones Biológicas, CSIC, Ramiro de Maeztu 9, E-28040 Madrid, Spain.