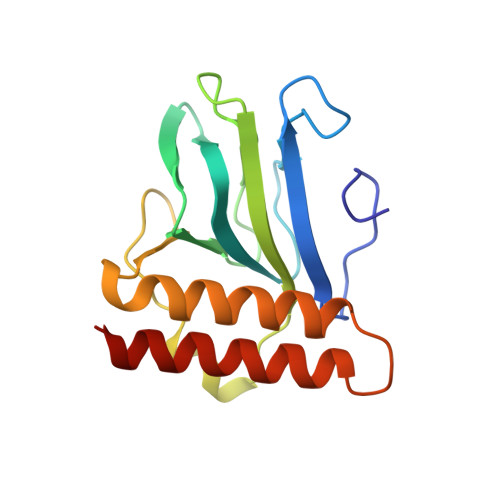
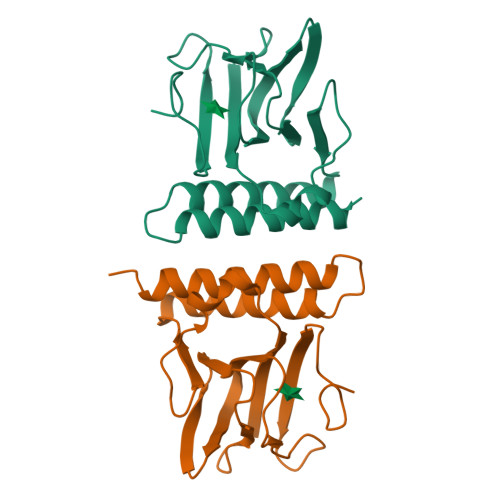Structural basis for ligand-regulated oligomerization of AraC.
Soisson, S.M., MacDougall-Shackleton, B., Schleif, R., Wolberger, C.(1997) Science 276: 421-425
- PubMed: 9103202
- DOI: https://doi.org/10.1126/science.276.5311.421
- Primary Citation of Related Structures:
2ARA, 2ARC - PubMed Abstract:
The crystal structure of the arabinose-binding and dimerization domain of the Escherchia coli gene regulatory protein AraC was determined in the presence and absence of L-arabinose. The 1.5 angstrom structure of the arabinose-bound molecule shows that the protein adopts an unusual fold, binding sugar within a beta barrel and completely burying the arabinose with the amino-terminal arm of the protein. Dimer contacts in the presence of arabinose are mediated by an antiparallel coiled-coil. In the 2.8 angstrom structure of the uncomplexed protein, the amino-terminal arm is disordered, uncovering the sugar-binding pocket and allowing it to serve as an oligomerization interface. The ligand-gated oligomerization as seen in AraC provides the basis of a plausible mechanism for modulating the protein's DNA-looping properties.
Organizational Affiliation:
Department of Biophysics and Biophysical Chemistry, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA.