Tryptophan 7-halogenase (PrnA) structure suggests a mechanism for regioselective chlorination.
Dong, C., Flecks, S., Unversucht, S., Haupt, C., van Pee, K.H., Naismith, J.H.(2005) Science 309: 2216-2219
- PubMed: 16195462
- DOI: https://doi.org/10.1126/science.1116510
- Primary Citation of Related Structures:
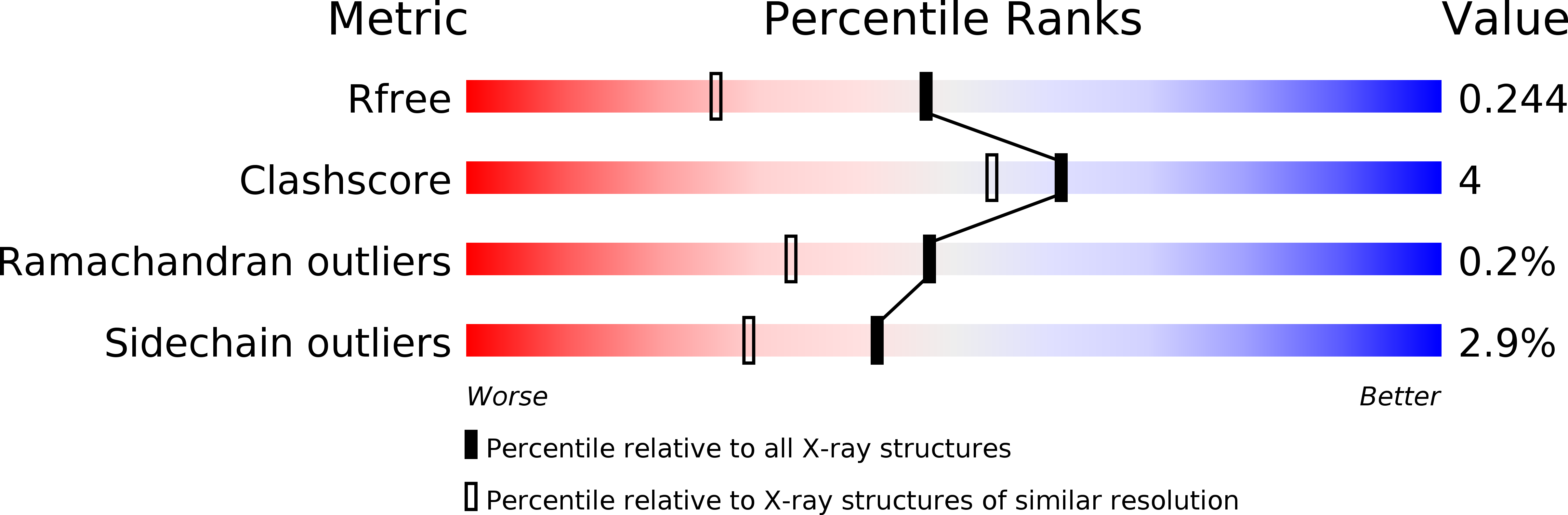
2APG, 2AQJ, 2AR8, 2ARD - PubMed Abstract:
Chlorinated natural products include vancomycin and cryptophycin A. Their biosynthesis involves regioselective chlorination by flavin-dependent halogenases. We report the structural characterization of tryptophan 7-halogenase (PrnA), which regioselectively chlorinates tryptophan. Tryptophan and flavin adenine dinucleotide (FAD) are separated by a 10 angstrom-long tunnel and bound by distinct enzyme modules. The FAD module is conserved in halogenases and is related to flavin-dependent monooxygenases. On the basis of biochemical studies, crystal structures, and by analogy with monooxygenases, we predict that FADH2 reacts with O2 to make peroxyflavin, which is decomposed by Cl-. The resulting HOCl is guided through the tunnel to tryptophan, where it is activated to participate in electrophilic aromatic substitution.
Organizational Affiliation:
Centre for Biomolecular Sciences, EaStchem, University of St. Andrews, St. Andrews KY16 9ST, UK.