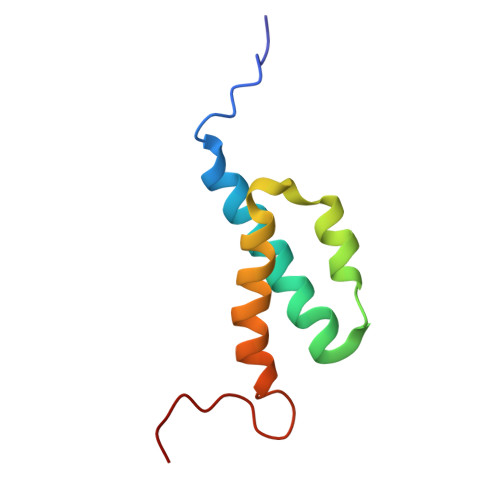
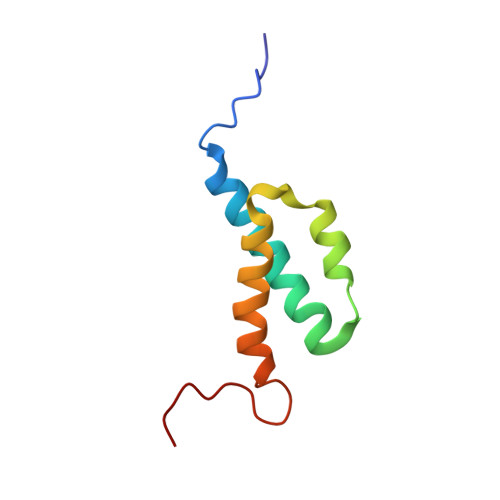
Nuclear Magnetic Resonance Solution Structure of the Escherichia coli DNA Polymerase III {theta} Subunit.
Mueller, G.A., Kirby, T.W., Derose, E.F., Li, D., Schaaper, R.M., London, R.E.(2005) J Bacteriol 187: 7081-7089
- PubMed: 16199579
- DOI: https://doi.org/10.1128/JB.187.20.7081-7089.2005
- Primary Citation of Related Structures:
2AE9 - PubMed Abstract:
The catalytic core of Escherichia coli DNA polymerase III holoenzyme contains three subunits: alpha, epsilon, and theta. The alpha subunit contains the polymerase, and the epsilon subunit contains the exonucleolytic proofreading function. The small (8-kDa) theta subunit binds only to epsilon. Its function is not well understood, although it was shown to exert a small stabilizing effect on the epsilon proofreading function. In order to help elucidate its function, we undertook a determination of its solution structure. In aqueous solution, theta yielded poor-quality nuclear magnetic resonance spectra, presumably due to conformational exchange and/or protein aggregation. Based on our recently determined structure of the theta homolog from bacteriophage P1, named HOT, we constructed a homology model of theta. This model suggested that the unfavorable behavior of theta might arise from exposed hydrophobic residues, particularly toward the end of alpha-helix 3. In gel filtration studies, theta elutes later than expected, indicating that aggregation is potentially responsible for these problems. To address this issue, we recorded 1H-15N heteronuclear single quantum correlation (HSQC) spectra in water-alcohol mixed solvents and observed substantially improved dispersion and uniformity of peak intensities, facilitating a structural determination under these conditions. The structure of theta in 60/40 (vol/vol) water-methanol is similar to that of HOT but differs significantly from a previously reported theta structure. The new theta structure is expected to provide additional insight into its physiological role and its effect on the epsilon proofreading subunit.
Organizational Affiliation:
Laboratory of Structural Biology, MR-01, National Institute of Environmental Health Sciences, 111 Alexander Drive, Research Triangle Park, North Carolina 27709, USA.