Structure of a liganded type 2 non-specific lipid-transfer protein from wheat and the molecular basis of lipid binding.
Hoh, F., Pons, J.L., Gautier, M.F., de Lamotte, F., Dumas, C.(2005) Acta Crystallogr D Biol Crystallogr 61: 397-406
- PubMed: 15805594
- DOI: https://doi.org/10.1107/S0907444905000417
- Primary Citation of Related Structures:
1TUK - PubMed Abstract:
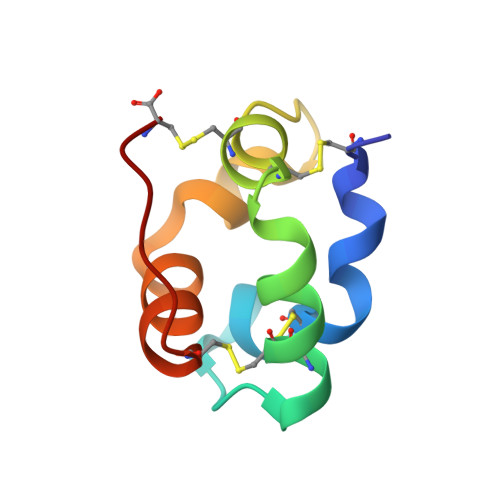
In plants, a family of ubiquitous proteins named non-specific lipid-transfer proteins (ns-LTPs) facilitates the transfer of fatty acids, phospholipids and steroids between membranes. Recent data suggest that these secreted proteins play a key role in the formation of cuticular wax layers and in defence mechanisms against pathogens. In this study, X-ray crystallography has been used to examine the structural details of the interaction between a wheat type 2 ns-LTP and a lipid, L-alpha-palmitoyl-phosphatidyl glycerol. This crystal structure was solved ab initio at 1.12 A resolution by direct methods. The typical alpha-helical bundle fold of this protein is maintained by four disulfide bridges and delineates two hydrophobic cavities. The inner surface of the main cavity is lined by non-polar residues that provide a hydrophobic environment for the palmitoyl moiety of the lipid. The head-group region of this lipid protrudes from the surface and makes several polar interactions with a conserved patch of basic residues at the entrance of the pocket. The alkyl chain of a second lipid is bound within an adjacent smaller cavity. The structure shows that binding of the lipid tails to the protein involves extensive hydrophobic interactions.
- Centre de Biochimie Structurale, CNRS UMR5048-INSERM U554, 29 Rue de Navacelles, 34090 Montpellier, France.
Organizational Affiliation: