Molecular Details of Urease Inhibition by Boric Acid: Insights into the Catalytic Mechanism.
Benini, S., Rypniewski, W.R., Wilson, K.S., Mangani, S., Ciurli, S.(2004) J Am Chem Soc 126: 3714-3715
- PubMed: 15038715
- DOI: https://doi.org/10.1021/ja049618p
- Primary Citation of Related Structures:
1S3T - PubMed Abstract:
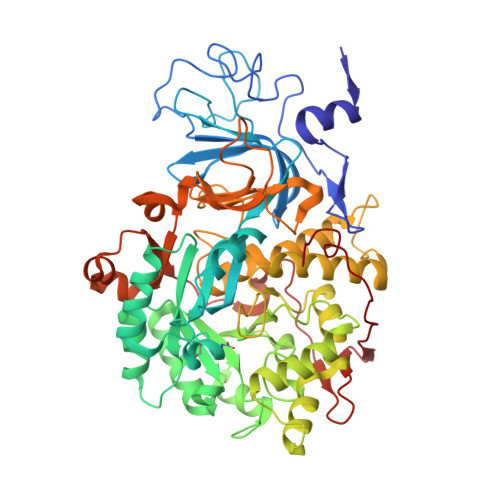
The structure of the complex of urease, a Ni-containing metalloenzyme, with boric acid was determined at 2.10 A resolution. The complex shows the unprecedented binding mode of the competitive inhibitor to the dinuclear metal center, with the B(OH)3 molecule bridging the Ni ions and leaving in place the bridging hydroxide. Boric acid can be considered a substrate analogue of urea, and the structure supports the proposal that the Ni-bridging hydroxide acts as the nucleophile in the enzymatic process of urea hydrolysis.
- York Structural Biology Laboratory, University of York, York, UK.
Organizational Affiliation: