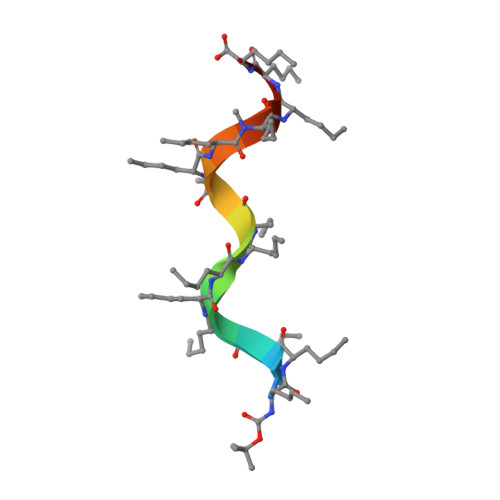
Solution structure of a D,L-alternating oligonorleucine as a model of double-stranded antiparallel beta-helix.
Navarro, E., Fenude, E., Celda, B.(2002) Biopolymers 64: 198-209
- PubMed: 12115137
- DOI: https://doi.org/10.1002/bip.10172
- Primary Citation of Related Structures:
1S1O - PubMed Abstract:
Conformational characteristics of alternating D,L linear peptides are of particular interest because of their capacity to form transmembrane channels with different transport properties, as some natural antibiotics do. Single- and double-stranded beta-helical structures are common for alternating D,L peptides. The stability of the beta-helix depends on several structural factors, such as the backbone peptide length, type and position of side chains, and nature of terminal groups. The NMR and molecular dynamics solution conformation of a synthetic alternating D,L-oligopeptide with 15 norleucines (XVMe) has been used as a model to get insight in to the conformational features of double-stranded beta-helix structures. The NH chemical shift values (delta(NH)) and long-range nuclear Overhauser effects (NOE) cross peaks, in particular interstrand connectivities, clearly point to an antiparallel double-stranded beta-helix for the XVMe major conformation in solution. An extensive set of distances (from NOE cross peaks) and H-bonds (from delta(NH)) has been included in the molecular dynamics calculations. The experimental NMR data and theoretical calculations clearly indicate that the most probable conformation of XVMe in solution is a double-strand antiparallel beta(5.6) increasing decreasing-helix structure.
- Departamento de Química Física, Facultat de Químicas, Universitat de Valencia, C/Dr. Moliner,50, 46100-Burjassot (Valencia), Spain.
Organizational Affiliation: