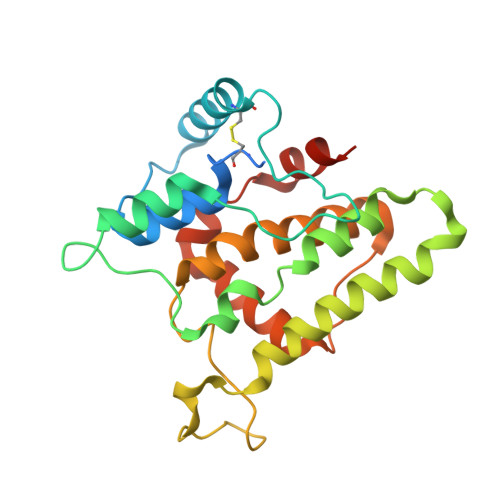
The Intracellular Chloride Ion Channel Protein CLIC1 Undergoes a Redox-controlled Structural Transition
Littler, D.R., Harrop, S.J., Fairlie, W.D., Brown, L.J., Pankhurst, G.J., Pankhurst, S., DeMaere, M.Z., Campbell, T.J., Bauskin, A.R., Tonini, R., Mazzanti, M., Breit, S.N., Curmi, P.M.(2004) J Biological Chem 279: 9298-9305
- PubMed: 14613939
- DOI: https://doi.org/10.1074/jbc.M308444200
- Primary Citation of Related Structures:
1RK4 - PubMed Abstract:
Most proteins adopt a well defined three-dimensional structure; however, it is increasingly recognized that some proteins can exist with at least two stable conformations. Recently, a class of intracellular chloride ion channel proteins (CLICs) has been shown to exist in both soluble and integral membrane forms. The structure of the soluble form of CLIC1 is typical of a soluble glutathione S-transferase superfamily protein but contains a glutaredoxin-like active site. In this study we show that on oxidation CLIC1 undergoes a reversible transition from a monomeric to a non-covalent dimeric state due to the formation of an intramolecular disulfide bond (Cys-24-Cys-59). We have determined the crystal structure of this oxidized state and show that a major structural transition has occurred, exposing a large hydrophobic surface, which forms the dimer interface. The oxidized CLIC1 dimer maintains its ability to form chloride ion channels in artificial bilayers and vesicles, whereas a reducing environment prevents the formation of ion channels by CLIC1. Mutational studies show that both Cys-24 and Cys-59 are required for channel activity.
- Initiative for Biomolecular Structure, School of Physics, University of New South Wales, Sydney, New South Wales 2052, Australia.
Organizational Affiliation: