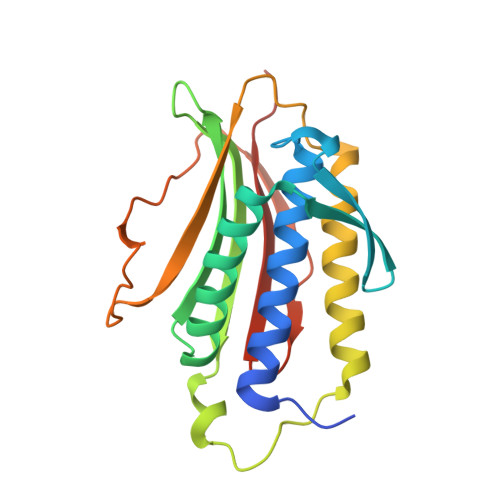
Crystal structure of the tetrameric Mad1-Mad2 core complex: implications of a 'safety belt' binding mechanism for the spindle checkpoint.
Sironi, L., Mapelli, M., Knapp, S., De Antoni, A., Jeang, K.T., Musacchio, A.(2002) EMBO J 21: 2496-2506
- PubMed: 12006501
- DOI: https://doi.org/10.1093/emboj/21.10.2496
- Primary Citation of Related Structures:
1GO4 - PubMed Abstract:
The spindle checkpoint protein Mad1 recruits Mad2 to unattached kinetochores and is essential for Mad2-Cdc20 complex formation in vivo but not in vitro. The crystal structure of the Mad1-Mad2 complex reveals an asymmetric tetramer, with elongated Mad1 monomers parting from a coiled-coil to form two connected sub-complexes with Mad2. The Mad2 C-terminal tails are hinged mobile elements wrapping around the elongated ligands like molecular 'safety belts'. We show that Mad1 is a competitive inhibitor of the Mad2-Cdc20 complex, and propose that the Mad1-Mad2 complex acts as a regulated gate to control Mad2 release for Cdc20 binding. Mad1-Mad2 is strongly stabilized in the tetramer, but a 1:1 Mad1-Mad2 complex slowly releases Mad2 for Cdc20 binding, driven by favourable binding energies. Thus, the rate of Mad2 binding to Cdc20 during checkpoint activation may be regulated by conformational changes that destabilize the tetrameric Mad1-Mad2 assembly to promote Mad2 release. We also show that unlocking the Mad2 C-terminal tail is required for ligand release from Mad2, and that the 'safety belt' mechanism may prolong the lifetime of Mad2-ligand complexes.
- Structural Biology Unit, Department of Experimental Oncology, European Institute of Oncology, Via Ripamonti 435, 20141 Milan, Italy.
Organizational Affiliation: