Kinetic and Structural Characterization of Phosphofructokinase from Lactobacillus bulgaricus.
Paricharttanakul, N.M., Ye, S., Menefee, A.L., Javid-Majd, F., Sacchettini, J.C., Reinhart, G.D.(2005) Biochemistry 44: 15280-15286
- PubMed: 16285731
- DOI: https://doi.org/10.1021/bi051283g
- Primary Citation of Related Structures:
1ZXX - PubMed Abstract:
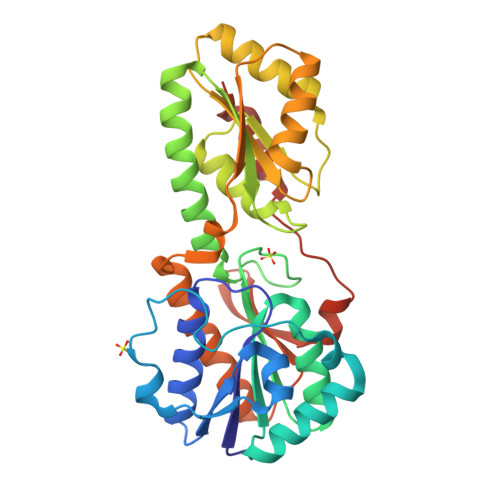
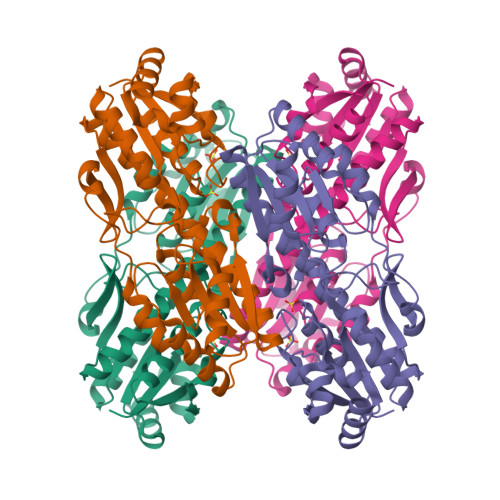
Phosphofructokinase from Lactobacillus delbrueckii subspecies bulgaricus (LbPFK) has been reported to be a nonallosteric analogue of phosphofructokinase from Escherichia coli at pH 8.2 [Le Bras et al. (1991) Eur. J. Biochem. 198, 683-687]. A reexamination of the kinetics of this enzyme shows LbPFK to have limited binding affinity toward the allosteric ligands, MgADP and PEP, with dissociation constants of approximately 20 mM for both. Their allosteric effects are observed only at high concentrations of these ligands, with both exhibiting inhibitory effects on substrate binding. No pH dependence was observed for the binding and the influence of MgADP and PEP on the enzyme. To attempt to explain these results, the crystal structure of LbPFK was solved using molecular replacement to 1.86 A resolution. A comparative study of the LbPFK structure with that of phosphofructokinases from E. coli (EcPFK) and Bacillus stearothermophilus (BsPFK) reveals a structure with conserved fold and substrate binding site. The effector binding site, however, shows many differences that could explain the observed decreases in binding affinity for MgADP and PEP in LbPFK as compared to the other two enzymes.
Organizational Affiliation:
Department of Biochemistry and Biophysics, Texas A&M University, and the Texas Agricultural Experiment Station, College Station, Texas 77843-2128, USA.