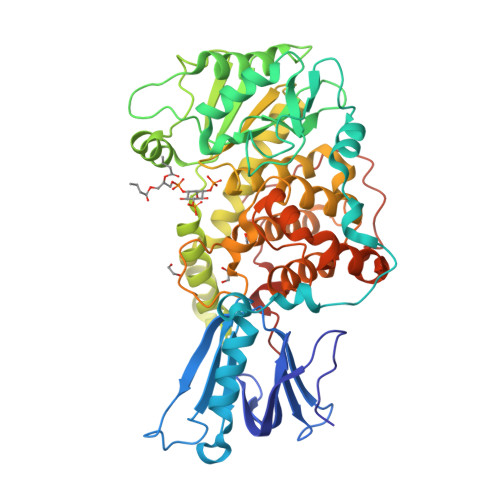
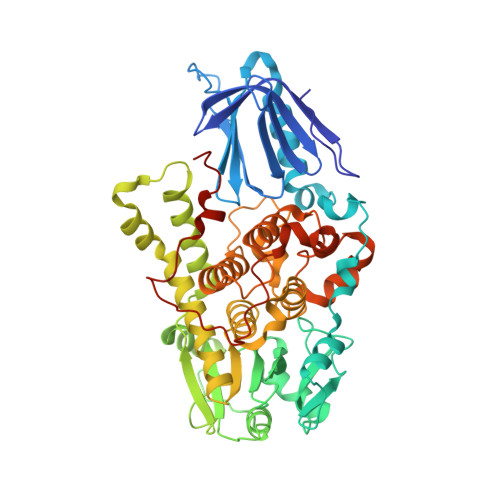
Molecular basis for substrate recognition by MTMR2, a myotubularin family phosphoinositide phosphatase
Begley, M.J., Taylor, G.S., Brock, M.A., Ghosh, P., Woods, V.L., Dixon, J.E.(2006) Proc Natl Acad Sci U S A 103: 927-932
- PubMed: 16410353
- DOI: https://doi.org/10.1073/pnas.0510006103
- Primary Citation of Related Structures:
1ZSQ, 1ZVR - PubMed Abstract:
Myotubularins, a large family of catalytically active and inactive proteins, belong to a unique subgroup of protein tyrosine phosphatases that use inositol phospholipids, rather than phosphoproteins, as physiological substrates. Here, by integrating crystallographic and deuterium-exchange mass spectrometry studies of human myotubularin-related protein-2 (MTMR2) in complex with phosphoinositides, we define the molecular basis for this unique substrate specificity. Phosphoinositide substrates bind in a pocket located on a positively charged face of the protein, suggesting an electrostatic mechanism for membrane targeting. A flexible, hydrophobic helix makes extensive interactions with the diacylglycerol moieties of substrates, explaining the specificity for membrane-bound phosphoinositides. An extensive H-bonding network and charge-charge interactions within the active site pocket determine phosphoinositide headgroup specificity. The conservation of these specificity determinants within the active, but not the inactive, myotubularins provides insight into the functional differences between the active and inactive members.
Organizational Affiliation:
Department of Pharmacology, University of California at San Diego, La Jolla, CA 92093, USA.