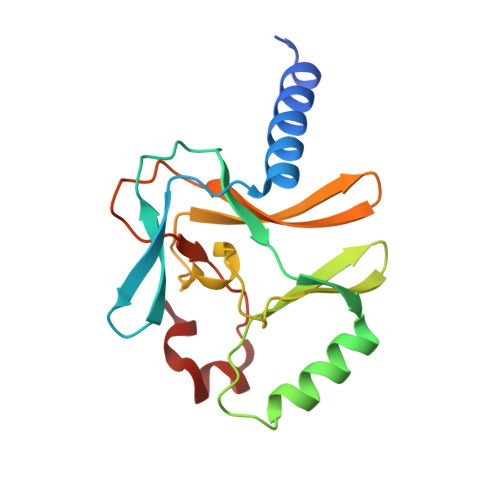
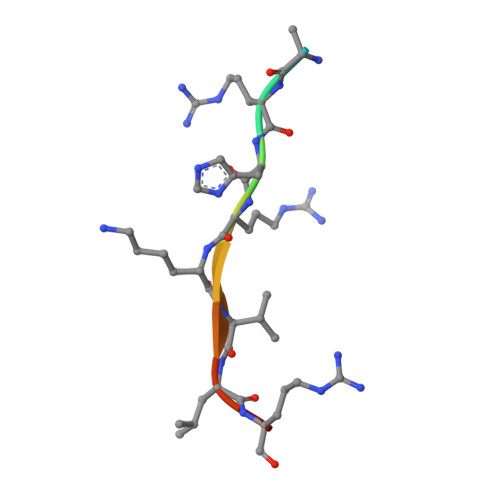
Structural and functional analysis of SET8, a histone H4 Lys-20 methyltransferase
Couture, J.-F., Collazo, E., Brunzelle, J.S., Trievel, R.C.(2005) Genes Dev 19: 1455-1465
- PubMed: 15933070
- DOI: https://doi.org/10.1101/gad.1318405
- Primary Citation of Related Structures:
1ZKK - PubMed Abstract:
SET8 (also known as PR-SET7) is a histone H4-Lys-20-specific methyltransferase that is implicated in cell-cycle-dependent transcriptional silencing and mitotic regulation in metazoans. Herein we report the crystal structure of human SET8 (hSET8) bound to a histone H4 peptide bearing Lys-20 and the product cofactor S-adenosylhomocysteine. Histone H4 intercalates in the substrate-binding cleft as an extended parallel beta-strand. Residues preceding Lys-20 in H4 engage in an extensive array of salt bridge, hydrogen bond, and van der Waals interactions with hSET8, while the C-terminal residues bind through predominantly hydrophobic interactions. Mutational analysis of both the substrate-binding cleft and histone H4 reveals that interactions with residues in the N and C termini of the H4 peptide are critical for conferring substrate specificity. Finally, analysis of the product specificity indicates that hSET8 is a monomethylase, consistent with its role in the maintenance of Lys-20 monomethylation during cell division.
Organizational Affiliation:
Department of Biological Chemistry, University of Michigan, Ann Arbor, Michigan 48109, USA.