Staphylococcus aureus Aminopeptidase S Is a Founding Member of a New Peptidase Clan.
Odintsov, S.G., Sabala, I., Bourenkov, G., Rybin, V., Bochtler, M.(2005) J Biol Chem 280: 27792-27799
- PubMed: 15932875
- DOI: https://doi.org/10.1074/jbc.M502023200
- Primary Citation of Related Structures:
1ZJC - PubMed Abstract:
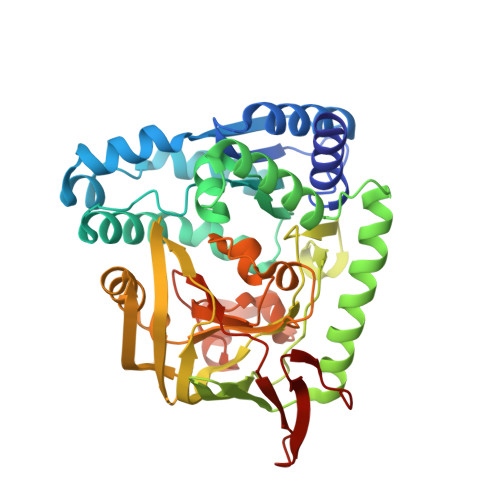
Staphylococcus aureus aminopeptidase S (AmpS) has been named for its predicted, but experimentally untested, aminopeptidase activity. The enzyme is homologous to biochemically characterized aminopeptidases that contain two cobalt or zinc ions in their active centers, but it is unrelated to all structurally characterized metallopeptidases. Here, we demonstrate AmpS aminopeptidase activity experimentally, and we present the 1.8-A crystal structure of the enzyme. Two metal ions with full occupancy and a third metal ion with low occupancy are present in the active site. A water molecule and Glu-319 serve as bridging ligands to the two metals with full occupancy. One of these metal ions is additionally coordinated by Glu-253 and His-348 and the other by His-381 and Asp-383. In addition, the metals are involved in weak metal-donor interactions to a water molecule and to Tyr-355. In the crystal, AmpS forms a dimer with a large internal cavity. The active sites are located at opposite ends of this internal cavity and are essentially inaccessible from the outside, suggesting that an inactive conformation was crystallized. Because gel filtration and analytical ultracentrifugation data also suggest dimer formation, the problem of substrate access to the active site cavity remains unresolved.
Organizational Affiliation:
International Institute of Molecular and Cell Biology, ul. Trojdena 4, 02-109 Warsaw, Poland.