A structural basis for CD8+ T cell-dependent recognition of non-homologous peptide ligands: implications for molecular mimicry in autoreactivity
Sandalova, T., Michaelsson, J., Harris, R.A., Odeberg, J., Schneider, G., Karre, K., Achour, A.(2005) J Biological Chem 280: 27069-27075
- PubMed: 15845547
- DOI: https://doi.org/10.1074/jbc.M500927200
- Primary Citation of Related Structures:
1ZHB - PubMed Abstract:
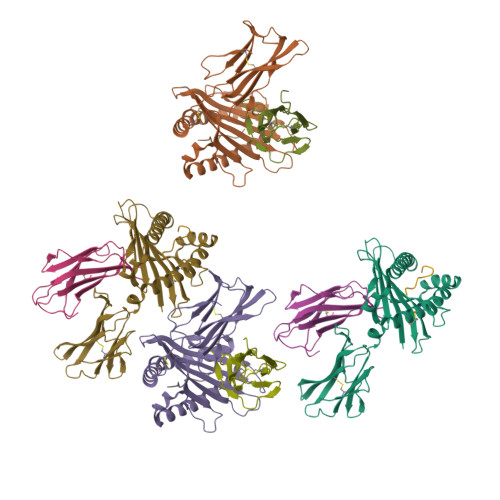
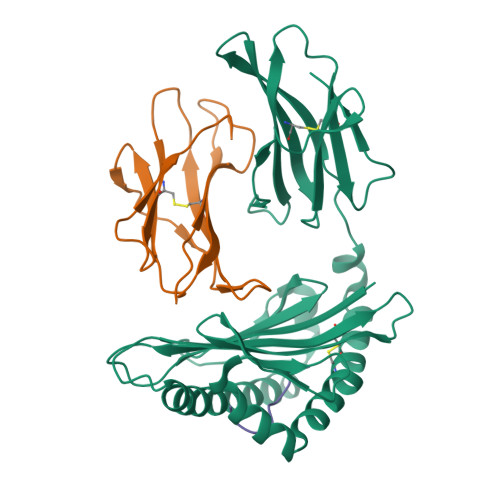
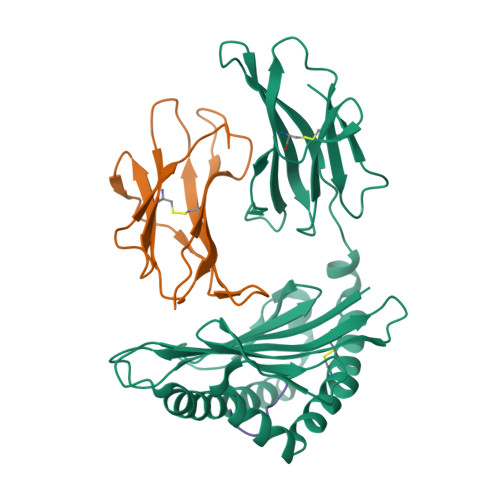
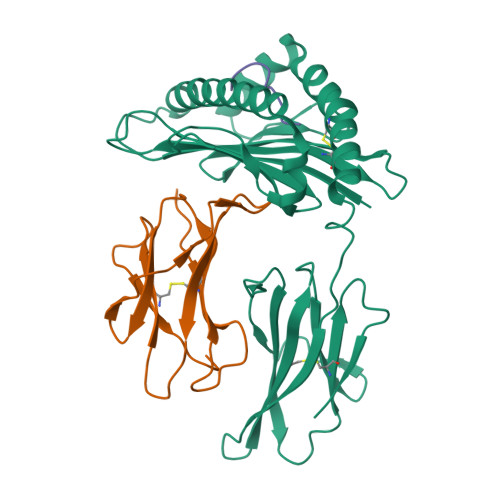
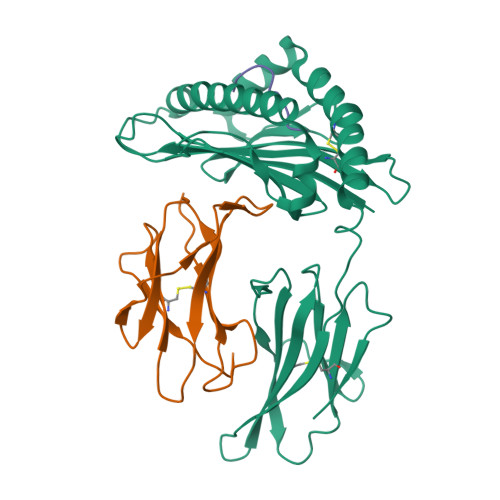
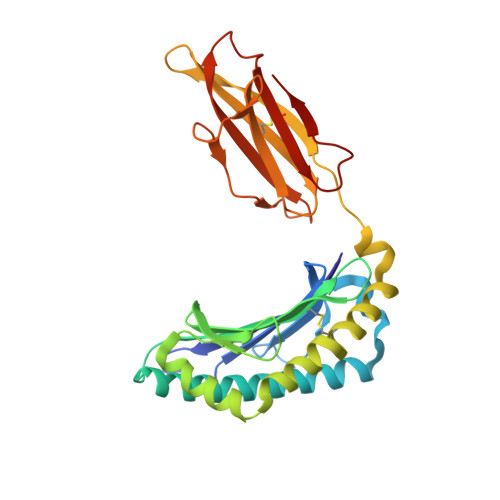
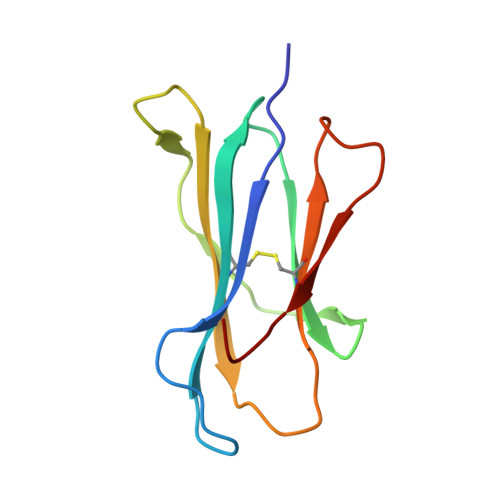
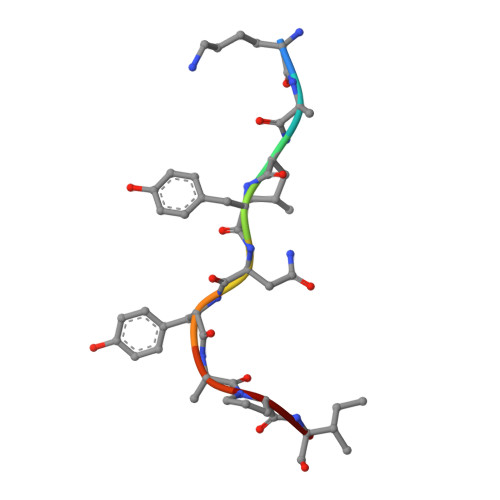
Molecular mimicry of self-epitopes by viral antigens is one possible pathogenic mechanism underlying induction of autoimmunity. A self-epitope, mDBM, derived from mouse dopamine beta-mono-oxygenase (KALYDYAPI) sharing 44% sequence identity with the lymphocytic choriomeningitis virus-derived immunodominant epitope gp33 (KAVYNFATC/M), has previously been identified as a cross-reactive self-ligand, presentation of which results in autoimmunity. A rat peptide homologue, rDBM (KALYNYAPI, 56% identity to gp33), which displayed similar properties to mDBM, has also been identified. We herein report the crystal structure of H-2Db.rDBM and a comparison with the crystal structures of the cross-reactive H-2Db.gp33 and non-cross-reactive H-2Db.gp33 (V3L) escape variant (KALYNFATM, 88% identity to gp33). Despite the large sequence disparity, rDBM and gp33 peptides are presented in nearly identical manners by H-2Db, with a striking juxtaposition of the central sections of both peptides from residues p3 to p7. The structural similarity provides H-2Db in complex with either a virus-derived or a dopamine beta-mono-oxygenase-derived peptide with a shared antigenic identity that conserves the positioning of the heavy chain and peptide residues that interact with the T cell receptor (TCR). This stands in contrast to the structure of H-2Db.gp33 (V3L), in which a single conserved mutation, also present in rDBM, induces large movements of both the peptide backbone and the side chains that interact with the TCR. The TCR-interacting surfaces of the H-2Db.rDBM and H-2Db.gp33 major histocompatibility complexes are very similar with regard to shape, topology, and charge distribution, providing a structural basis for CD8 T cell activation by molecular mimicry and potential subsequent development of autoreactivity.
Organizational Affiliation:
Department of Medical Biochemistry and Biophysics, Microbiology and Tumor Biology Center, and Strategic Research Center IRIS for Studies of Integrated Recognition in the Immune System, Karolinska Institutet, SE-171 77 Stockholm, Sweden.