Pathological crystallography: case studies of several unusual macromolecular crystals.
Dauter, Z., Botos, I., LaRonde-LeBlanc, N., Wlodawer, A.(2005) Acta Crystallogr D Biol Crystallogr 61: 967-975
- PubMed: 15983420
- DOI: https://doi.org/10.1107/S0907444905011285
- Primary Citation of Related Structures:
1Z0T, 1Z0V - PubMed Abstract:
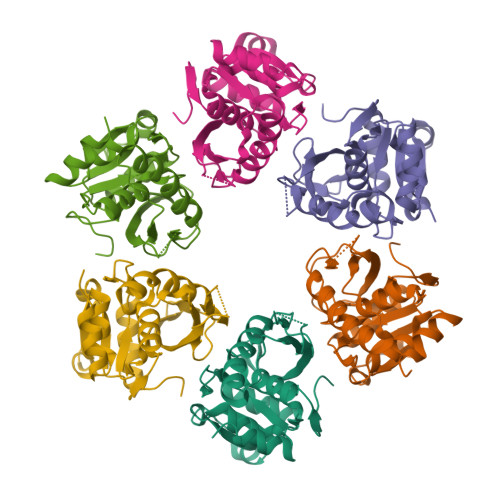
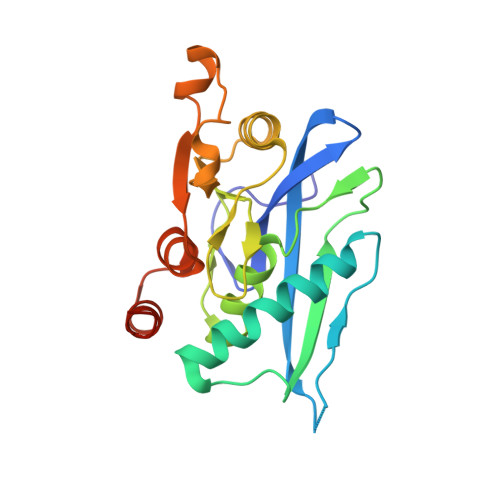
Although macromolecular crystallography is rapidly becoming largely routine owing to advances in methods of data collection, structure solution and refinement, difficult cases are still common. To remind structural biologists about the kinds of crystallographic difficulties that might be encountered, case studies of several successfully completed structure determinations that utilized less than perfect crystals are discussed here. The structure of the proteolytic domain of Archaeoglobus fulgidus Lon was solved with crystals that contained superimposed orthorhombic and monoclinic lattices, a case not previously described for proteins. Another hexagonal crystal form of this protein exhibited an unusually high degree of non-isomorphism. Crystals of A. fulgidus Rio1 kinase exhibited both pseudosymmetry and twinning. Ways of identifying the observed phenomena and approaches to solving and refining macromolecular structures when only less than perfect crystals are available are discussed here.
Organizational Affiliation:
Synchrotron Radiation Research Section, Macromolecular Crystallography Laboratory, National Cancer Institute and Biosciences Division, Argonne National Laboratory, Argonne, IL 60439, USA.