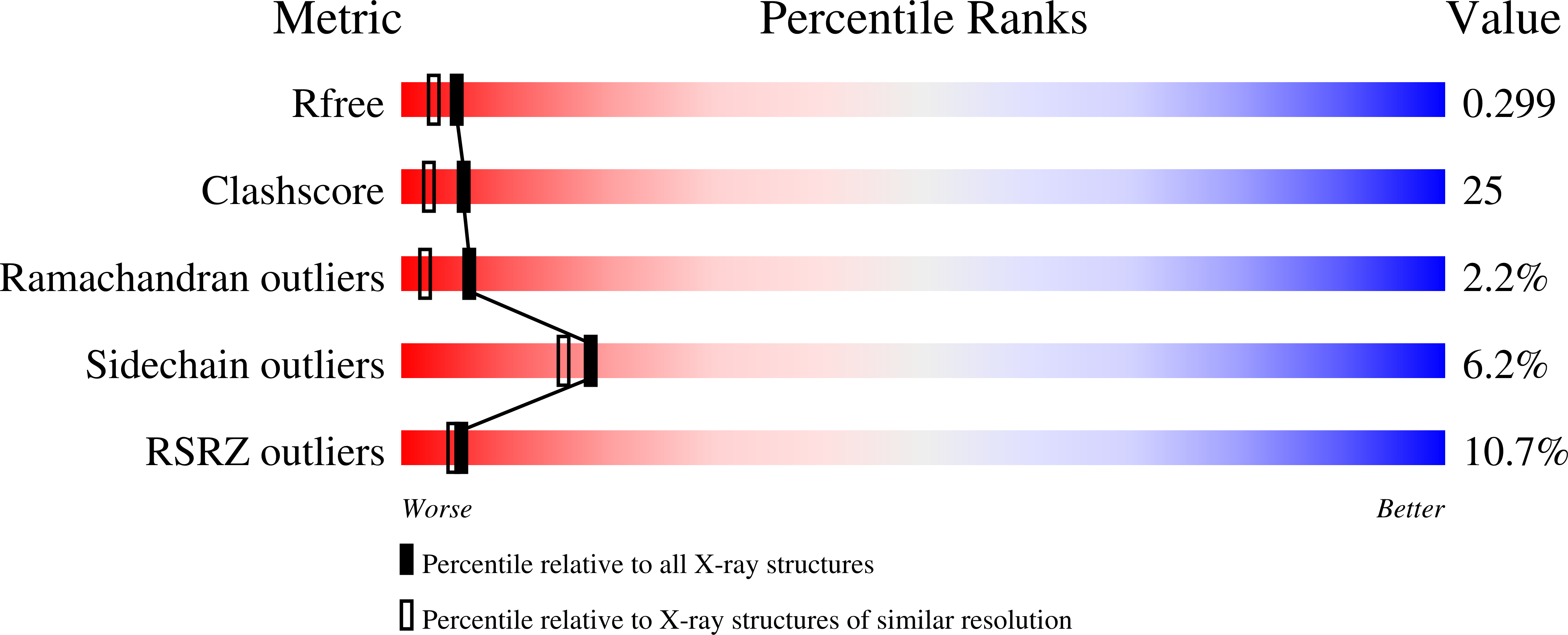
The Channel Architecture of Aquaporin 0 at a 2.2-A Resolution
Harries, W.E.C., Akhavan, D., Miercke, L.J.W., Khademi, S., Stroud, R.M.(2004) Proc Natl Acad Sci U S A 101: 14045-14050
- PubMed: 15377788
- DOI: https://doi.org/10.1073/pnas.0405274101
- Primary Citation of Related Structures:
1YMG - PubMed Abstract:
We determined the x-ray structure of bovine aquaporin 0 (AQP0) to a resolution of 2.2 A. The structure of this eukaryotic, integral membrane protein suggests that the selectivity of AQP0 for water transport is based on the identity and location of signature amino acid residues that are hallmarks of the water-selective arm of the AQP family of proteins. Furthermore, the channel lumen is narrowed only by two, quasi-2-fold related tyrosine side chains that might account for reduced water conductance relative to other AQPs. The channel is functionally open to the passage of water because there are eight discreet water molecules within the channel. Comparison of this structure with the recent electron-diffraction structure of the junctional form of sheep AQP0 at pH 6.0 that was interpreted as closed shows no global change in the structure of AQP0 and only small changes in side-chain positions. We observed no structural change to the channel or the molecule as a whole at pH 10, which could be interpreted as the postulated pH-gating mechanism of AQP0-mediated water transport at pH >6.5. Contrary to the electron-diffraction structure, the comparison shows no evidence of channel gating induced by association of the extracellular domains of AQP0 at pH 6.0. Our structure aids the analysis of the interaction of the extracellular domains and the possibility of a cell-cell adhesion role for AQP0. In addition, our structure illustrates the basis for formation of certain types of cataracts that are the result of mutations.
Organizational Affiliation:
Macromolecular Structure Group, Department of Biochemistry and Biophysics, University of California, S-412C Genentech Hall, 600 16th Street, San Francisco, CA 94143-2240, USA.