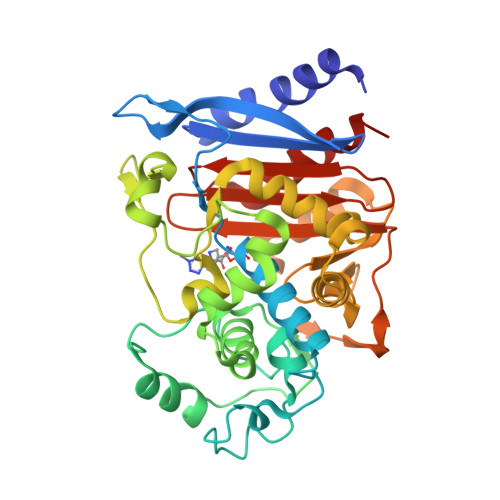
Crystal Structure of BRL 42715, C6-(N1-Methyl-1,2,3-triazolylmethylene)penem, in Complex with Enterobactercloacae 908R beta-Lactamase: Evidence for a Stereoselective Mechanism from Docking Studies
Michaux, C., Charlier, P., Frere, J.-M., Wouters, J.(2005) J Am Chem Soc 127: 3262-3263
- PubMed: 15755127
- DOI: https://doi.org/10.1021/ja0426241
- Primary Citation of Related Structures:
1Y54 - PubMed Abstract:
BRL 42715, C6-(N1-methyl-1,2,3-triazolylmethylene)penem, is an active-site-directed inactivator of bacterial beta-lactamases. The crystal structure of Enterobacter cloacae 908R class C beta-lactamase in complex with BRL 42715, docking, and energy minimization studies explain stereoselectivity of the binding of C6-(heterocyclic methylene)penems against class C beta-lactamase.
Organizational Affiliation:
Laboratoire de Chimie Biologique Structurale, Université of Namur, 5000 Namur, Belgium.