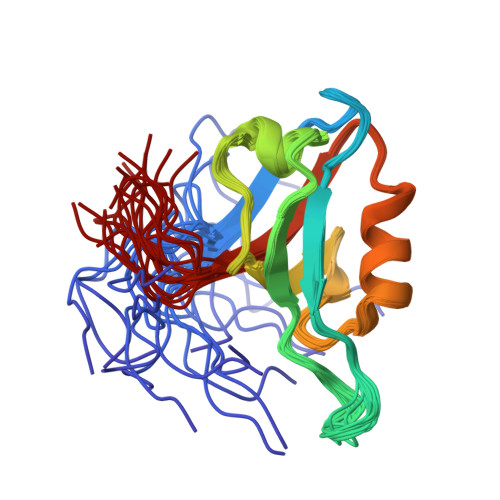
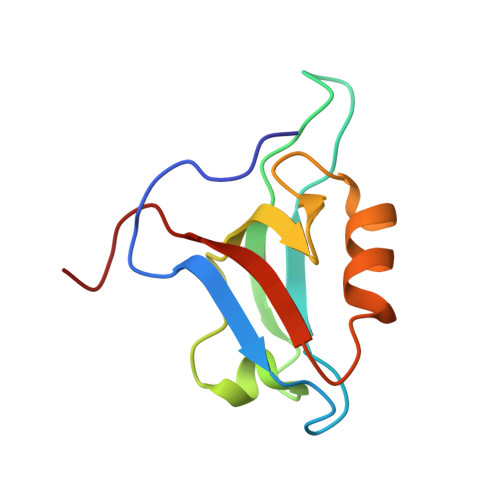
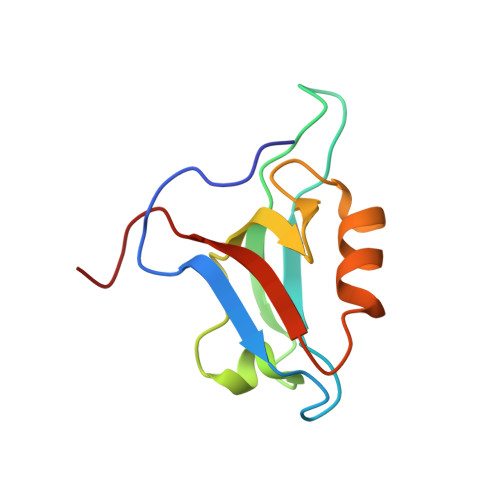
Discovery of low-molecular-weight ligands for the AF6 PDZ domain.
Joshi, M., Vargas, C., Boisguerin, P., Diehl, A., Krause, G., Schmieder, P., Moelling, K., Hagen, V., Schade, M., Oschkinat, H.(2006) Angew Chem Int Ed Engl 45: 3790-3795
- PubMed: 16671149
- DOI: https://doi.org/10.1002/anie.200503965
- Primary Citation of Related Structures:
1XZ9, 2EXG
Organizational Affiliation:
Leibniz-Institut für Molekulare Pharmakologie FMP, Robert-Roessle-Strasse 10, 13125 Berlin, Germany.