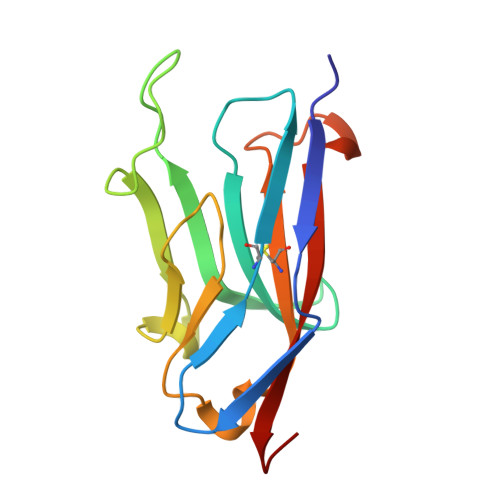
Ancient evolutionary origin of diversified variable regions demonstrated by crystal structures of an immune-type receptor in amphioxus.
Haire, R.N., Allaire, M., Jakoncic, J., Stojanoff, V., Cannon, J.P., Litman, G.W., Ostrov, D.A.(2006) Nat Immunol 7: 875-882
- PubMed: 16799561
- DOI: https://doi.org/10.1038/ni1359
- Primary Citation of Related Structures:
1XT5, 2FBO - PubMed Abstract:
Although the origins of genes encoding the rearranging binding receptors remain obscure, it is predicted that their ancestral forms were nonrearranging immunoglobulin-type domains. Variable region-containing chitin-binding proteins (VCBPs) are diversified immune-type molecules found in amphioxus (Branchiostoma floridae), an invertebrate that diverged early in deuterostome phylogeny. To study the potential evolutionary relationships between VCBPs and vertebrate adaptive immune receptors, we solved the structures of both a single V-type domain (to 1.15 A) and a pair of V-type domains (to 1.85 A) from VCBP3. The deduced structures show integral features of the ancestral variable-region fold as well as unique features of variable-region pairing in molecules that may reflect characteristics of ancestral forms of diversified immune receptors found in modern-day vertebrates.
Organizational Affiliation:
Department of Pathology, Immunology and Laboratory Medicine, University of Florida College of Medicine, Gainesville, Florida 32610, USA.