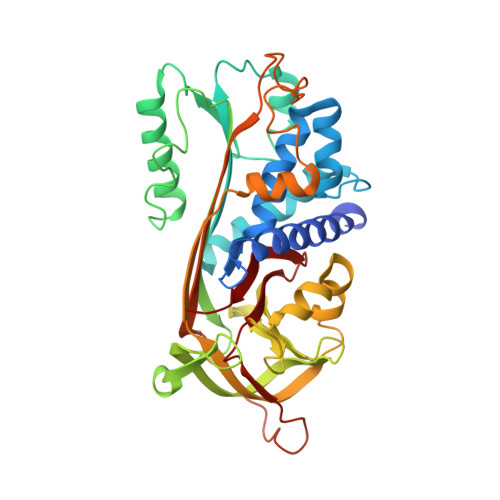
Crystal Structure of Human Maspin, a Serpin with Antitumor Properties: Reactive Center Loop of Mapsin is Exposed but Constrained
Al-Ayyoubi, M., Gettins, P.G., Volz, K.(2004) J Biol Chem 279: 55540-55544
- PubMed: 15501821
- DOI: https://doi.org/10.1074/jbc.M409957200
- Primary Citation of Related Structures:
1XQG, 1XQJ - PubMed Abstract:
Maspin, a member of the serpin superfamily, has tumor suppressing activity against breast and prostate cancer. Maspin inhibits tumor growth by blocking cell invasion, and its reactive center loop (RCL) is thought to mediate this activity. To understand this function on the molecular level, we have solved the three-dimensional structure of Maspin to 3.1 A resolution. The molecular structure shows the characteristic features of the serpin fold, but the RCL of Maspin is unique in length, composition, and placement. Although the RCL of Maspin is accessible and cleavable by some proteinases, it functions in the uncleaved, constrained conformation observed here. These structural results will contribute to our understanding of the mechanism by which Maspin suppresses tumors.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, University of Illinois at Chicago, 835 S. Wolcott Avenue, Chicago, IL 60612-7334.