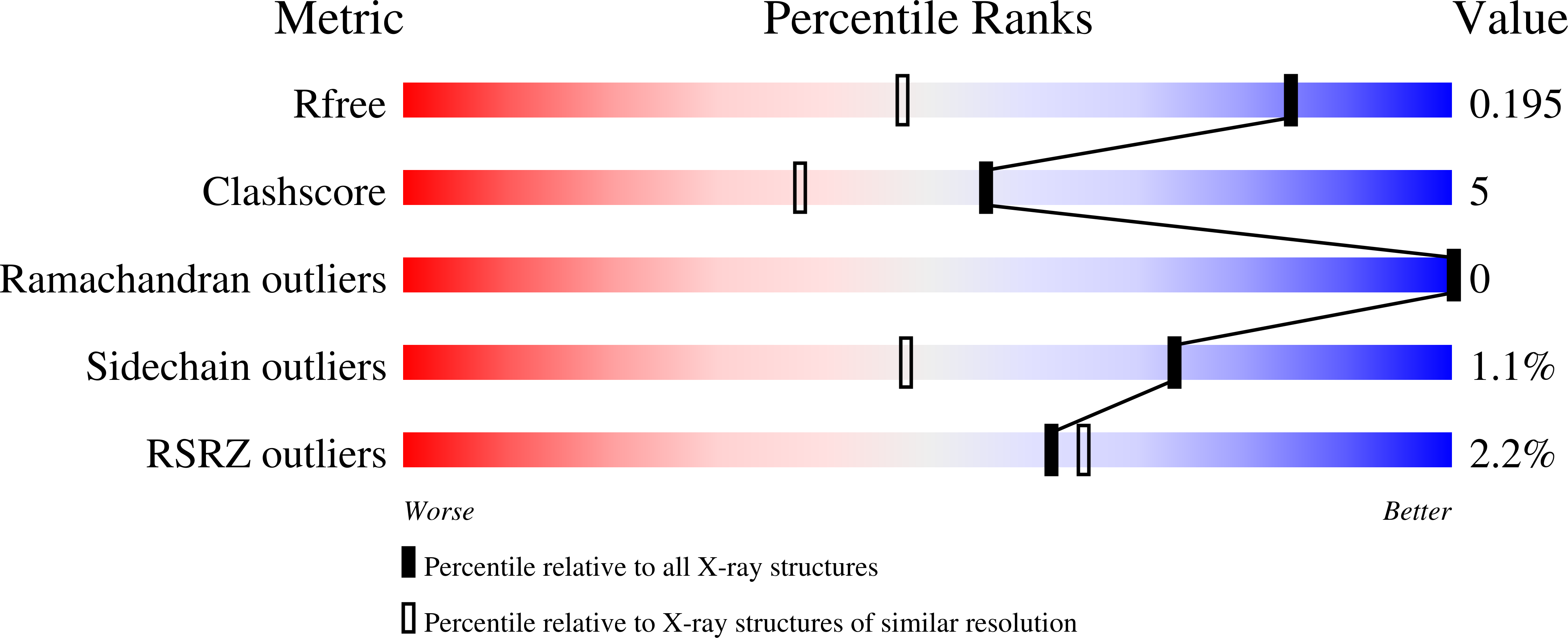
Redox-dependent Structural Reorganization in Putidaredoxin, a Vertebrate-type [2Fe-2S] Ferredoxin from Pseudomonas putida.
Sevrioukova, I.F.(2005) J Mol Biol 347: 607-621
- PubMed: 15755454
- DOI: https://doi.org/10.1016/j.jmb.2005.01.047
- Primary Citation of Related Structures:
1XLN, 1XLO, 1XLP, 1XLQ - PubMed Abstract:
Putidaredoxin (Pdx), a vertebrate-type [2Fe-2S] ferredoxin from Pseudomonas putida, transfers electrons from NADH-putidaredoxin reductase to cytochrome P450cam. Pdx exhibits redox-dependent binding affinities for P450cam and is thought to play an effector role in the monooxygenase reaction catalyzed by this hemoprotein. To understand how the reduced form of Pdx is stabilized and how reduction of the [2Fe-2S] cluster affects molecular properties of the iron-sulfur protein, crystal structures of reduced C73S and C73S/C85S Pdx were solved to 1.45 angstroms and 1.84 angstroms resolution, respectively, and compared to the corresponding 2.0 angstroms and 2.03 angstroms X-ray models of the oxidized mutants. To prevent photoreduction, the latter models were determined using in-house radiation source and the X-ray dose received by Pdx crystals was significantly decreased. Structural analysis showed that in reduced Pdx the Cys45-Ala46 peptide bond flip initiates readjustment of hydrogen bonding interactions between the [2Fe-2S] cluster, the Sgamma atoms of the cysteinyl ligands, and the backbone amide nitrogen atoms that results in tightening of the Cys39-Cys48 metal cluster binding loop around the prosthetic group and shifting of the metal center toward the Cys45-Thr47 peptide. From the metal center binding loop, the redox changes are transmitted to the linked Ile32-Asp38 peptide triggering structural rearrangement between the Tyr33-Asp34, Ser7-Asp9 and Pro102-Asp103 fragments of Pdx. The newly established hydrogen bonding interactions between Ser7, Asp9, Tyr33, Asp34, and Pro102, in turn, not only stabilize the tightened conformation of the [2Fe-2S] cluster binding loop but also assist in formation of a specific structural patch on the surface of Pdx that can be recognized by P450cam. This redox-linked change in surface properties is likely to be responsible for different binding affinity of oxidized and reduced Pdx to the hemoprotein.
Organizational Affiliation:
Department of Molecular Biology and Biochemistry, University of California, Irvine, CA 92612-3900, USA. sevrioui@uci.edu