Molecular properties of two proteins homologous to PduO-type ATP:cob(I)alamin adenosyltransferase from Sulfolobus tokodaii
Tanaka, Y., Sasaki, T., Kumagai, I., Yasutake, Y., Yao, M., Tanaka, I., Tsumoto, K.(2007) Proteins 68: 446-457
- PubMed: 17492665
- DOI: https://doi.org/10.1002/prot.21303
- Primary Citation of Related Structures:
1WVT - PubMed Abstract:
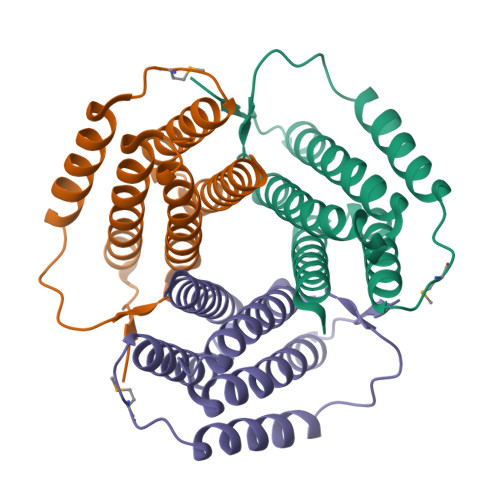
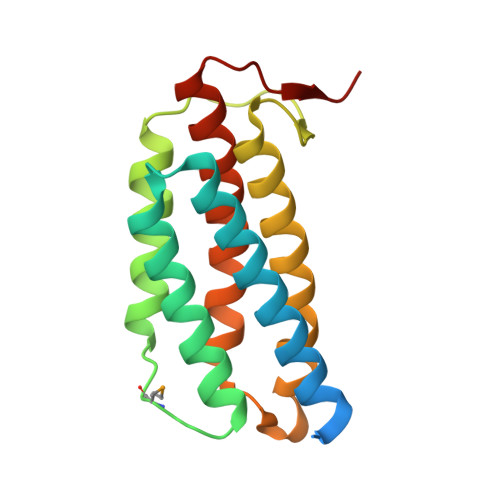
In the thermophilic archaeon Sulfolobus tokodaii, there are two genes homologous to PduO-type ATP:cob(I)alamin adenosyltransferase, ST1454 and ST2180. To address the structure and function of these two sequence-related proteins from one organism, we prepared them by using the Escherichia coli expression system and analyzed them by immunoblotting, matrix-assisted laser desorption ionization-time-of-flight mass spectroscopy, circular dichroism spectrometry, ATP:cobalamin adenosyltransferase assay, and X-ray crystallography. Immunoblotting and matrix-assisted laser desorption ionization-time-of-flight mass spectroscopy analyses showed that both these proteins are expressed in S. tokodaii cells as soluble proteins and are spontaneously digested at the N-terminal region. ATP:cob(I)alamin adenosyltransferase activity was detected for ST1454 but not for ST2180. ST2180 reduced the concentration of cob(I)alamin, suggesting that ST2180 might recognize cob(I)alamin as a ligand. The secondary structure of ST1454 was retained even in 7 M guanidine hydrochroride, whereas that of ST2180 was melted in 4.5 M guanidine hydrochloride. The X-ray crystal structural analysis revealed that the proteins shared a common structure: a trimer of five-helix bundles with a clockwise kink. There is a pocket surrounded by highly conserved residues, in which a polypropylene glycol 400 in the crystal structure of ST1454 was captured, suggesting that it is an active site. Structural comparison between these two proteins showed the difference in the number of ion pairs around the proposed active site. On the basis of these results, we propose that ST1454 and ST2180 have related but distinct functions.
Organizational Affiliation:
Department of Biomolecular Engineering, Graduate School of Engineering, Tohoku University, Sendai, Japan.