ATP Increases the Affinity between Muts ATPase Domains: Implications for ATP Hydrolysis and Conformational Changes
Lamers, M.H., Georgijevic, D., Lebbink, J., Winterwerp, H.H.K., Agianian, B., De Wind, N., Sixma, T.K.(2004) J Biological Chem 279: 43879
- PubMed: 15297450
- DOI: https://doi.org/10.1074/jbc.M406380200
- Primary Citation of Related Structures:
1W7A - PubMed Abstract:
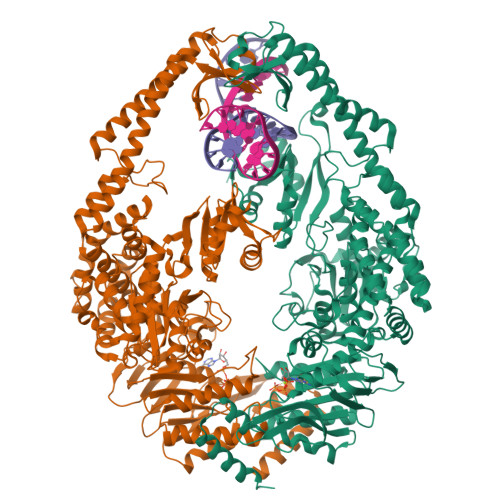
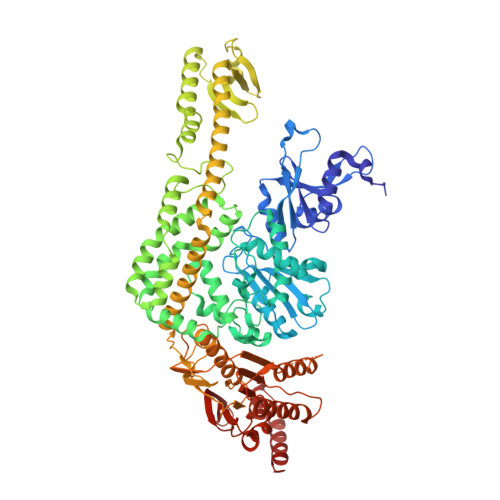
MutS is the key protein of the Escherichia coli DNA mismatch repair system. It recognizes mispaired and unpaired bases and has intrinsic ATPase activity. ATP binding after mismatch recognition by MutS serves as a switch that enables MutL binding and the subsequent initiation of mismatch repair. However, the mechanism of this switch is poorly understood. We have investigated the effects of ATP binding on the MutS structure. Crystallographic studies of ATP-soaked crystals of MutS show a trapped intermediate, with ATP in the nucleotide-binding site. Local rearrangements of several residues around the nucleotide-binding site suggest a movement of the two ATPase domains of the MutS dimer toward each other. Analytical ultracentrifugation experiments confirm such a rearrangement, showing increased affinity between the ATPase domains upon ATP binding and decreased affinity in the presence of ADP. Mutations of specific residues in the nucleotide-binding domain reduce the dimer affinity of the ATPase domains. In addition, ATP-induced release of DNA is strongly reduced in these mutants, suggesting that the two activities are coupled. Hence, it seems plausible that modulation of the affinity between ATPase domains is the driving force for conformational changes in the MutS dimer. These changes are driven by distinct amino acids in the nucleotide-binding site and form the basis for long-range interactions between the ATPase domains and DNA-binding domains and subsequent binding of MutL and initiation of mismatch repair.
Organizational Affiliation:
Division of Molecular Carcinogenesis, Netherlands Cancer Institute, Plesmanlaan 121, 1066 CX Amsterdam.