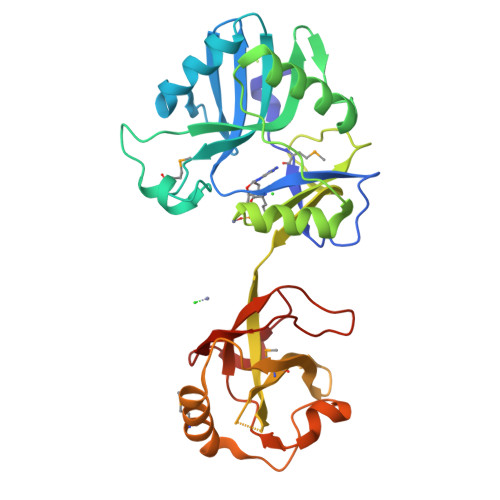
Crystal Structure and Nonhomologous End-joining Function of the Ligase Component of Mycobacterium DNA Ligase D.
Akey, D., Martins, A., Aniukwu, J., Glickman, M.S., Shuman, S., Berger, J.M.(2006) J Biological Chem 281: 13412-13423
- PubMed: 16476729
- DOI: https://doi.org/10.1074/jbc.M513550200
- Primary Citation of Related Structures:
1VS0 - PubMed Abstract:
DNA ligase D (LigD) is a large polyfunctional enzyme involved in nonhomologous end-joining (NHEJ) in mycobacteria. LigD consists of a C-terminal ATP-dependent ligase domain fused to upstream polymerase and phosphoesterase modules. Here we report the 2.4 angstroms crystal structure of the ligase domain of Mycobacterium LigD, captured as the covalent ligase-AMP intermediate with a divalent metal in the active site. A chloride anion on the protein surface coordinated by the ribose 3'-OH and caged by arginine and lysine side chains is a putative mimetic of the 5'-phosphate at a DNA nick. Structure-guided mutational analysis revealed distinct requirements for the adenylylation and end-sealing reactions catalyzed by LigD. We found that a mutation of Mycobacterium LigD that ablates only ligase activity results in decreased fidelity of NHEJ in vivo and a strong bias of mutagenic events toward deletions instead of insertions at the sealed DNA ends. This phenotype contrasts with the increased fidelity of double-strand break repair in deltaligD cells or in a strain in which only the polymerase function of LigD is defective. We surmise that the signature error-prone quality of bacterial NHEJ in vivo arises from a dynamic balance between the end-remodeling and end-sealing steps.
Organizational Affiliation:
Department of Molecular and Cellular Biology, University of California, Berkeley, California 94720.