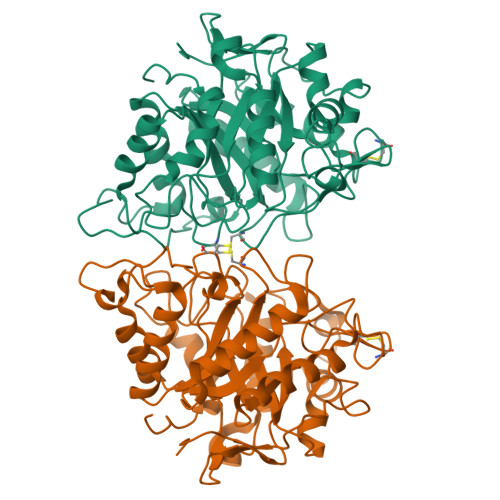
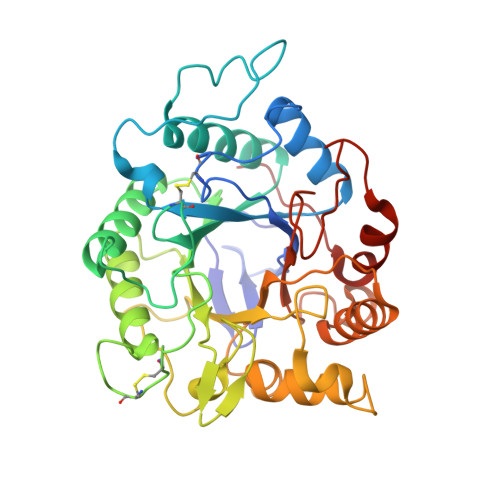
Catalytically enhanced endocellulase Cel5A from Acidothermus cellulolyticus.
Baker, J.O.,
McCarley, J.R.,
Lovett, R.,
Yu, C.H.,
Adney, W.S.,
Rignall, T.R.,
Vinzant, T.B.,
Decker, S.R.,
Sakon, J.,
Himmel, M.E.(2005) Appl Biochem Biotechnol 121-124: 129-148
- PubMed: 15917594 Search on PubMed
- Primary Citation of Related Structures:
1VRX
- PubMed Abstract:
Organizational Affiliation: National Bioenergy Center, National Renewable Energy Laboratory, 1617 Cole Boulevard, Golden, CO 80401, USA. john_baker@nrel.gov