Proton nuclear magnetic resonance and distance geometry/simulated annealing studies on the variant-1 neurotoxin from the New World scorpion Centruroides sculpturatus Ewing.
Lee, W., Jablonsky, M.J., Watt, D.D., Krishna, N.R.(1994) Biochemistry 33: 2468-2475
- PubMed: 8117707
- DOI: https://doi.org/10.1021/bi00175a015
- Primary Citation of Related Structures:
1VNA, 1VNB - PubMed Abstract:
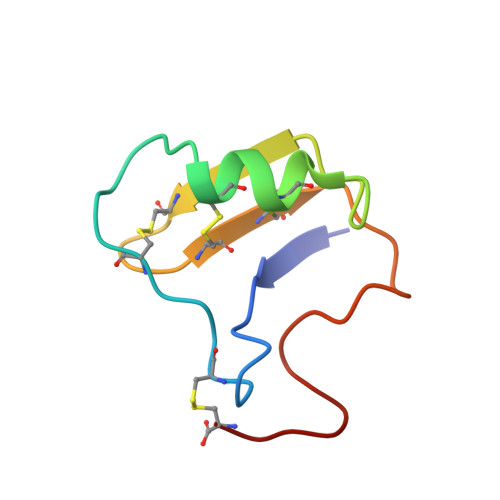
The sequence-specific proton resonance assignments for the variant-1 (CsE-v1) neurotoxin from the venom of the New World scorpion Centruroides sculpturatus Ewing (range Southwestern United States) have been performed by 2D 1H NMR spectroscopy at 600 MHz. The stereospecific assignments for the beta-methylene protons of 19 non-proline residues have been determined. A number of short-, medium-, and long-range NOESY contacts as well as the backbone and the side-chain vicinal coupling constants for several residues have been determined. Slowly exchanging amide hydrogens from a number of residues have been identified. On the basis of the NMR data, the solution structure of this protein has been determined by a hybrid procedure consisting of distance geometry and dynamical simulated annealing refinement calculations. Distance constraints from the NOESY data and torsion angle constraints from proton vicinal coupling constant data were used in the simulated annealing calculations. The three-dimensional structure of CsE-v1 is characterized by a three-stranded antiparallel beta-sheet, a short alpha-helix, a cis-proline, and intervening loops. A comparison with the solution NMR data of a homologous protein (CsE-v3) from the Centruroides venom, shows that the structures are essentially similar, except for some minor differences. Some of the NMR spectral perturbations are felt in regions far removed from sites of amino acid substitutions. The hydrophobic surface in CsE-v1 is slightly more extended than in CsE-v3.
Organizational Affiliation:
Department of Biochemistry, University of Alabama at Birmingham 35294.