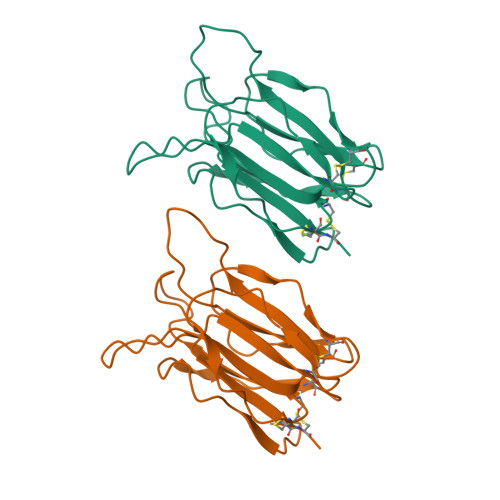
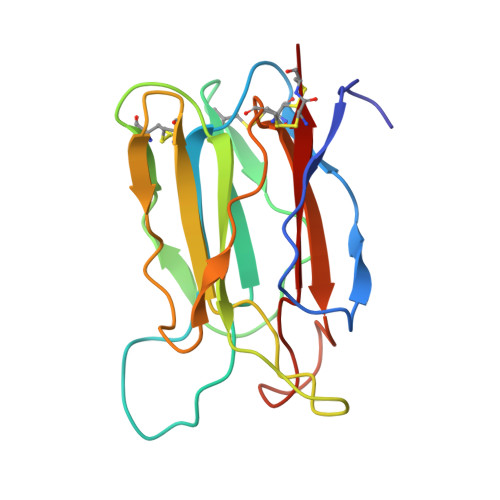
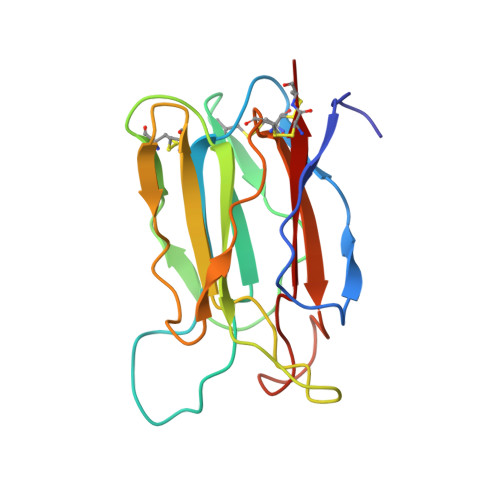
Crystal structure of vitelline membrane outer layer protein I (VMO-I): a folding motif with homologous Greek key structures related by an internal three-fold symmetry.
Shimizu, T., Vassylyev, D.G., Kido, S., Doi, Y., Morikawa, K.(1994) EMBO J 13: 1003-1010
- PubMed: 8131734
- DOI: https://doi.org/10.1002/j.1460-2075.1994.tb06348.x
- Primary Citation of Related Structures:
1VMO - PubMed Abstract:
The crystal structure of vitelline membrane outer layer protein I (VMO-I), which is isolated from the vitelline membrane outer layer of hen's eggs, has been determined by the multiple isomorphous replacement method and refined to an R-factor of 18.8% at 2.2 A resolution. The main chain folds into an unusual structure that consists of three beta-sheets forming Greek key motifs, which are related by an internal pseudo three-fold symmetry. The internal portion surrounded by these three beta-sheets is filled with hydrophobic side chains. This conformational feature coincides with three internal repeats in the sequence. Although a similar fold exists in the second domain of delta-endotoxin, there are significant structural differences between the two proteins, with the three-fold symmetry being most regular in VMO-I.
Organizational Affiliation:
Protein Engineering Research Institute, Osaka, Japan.