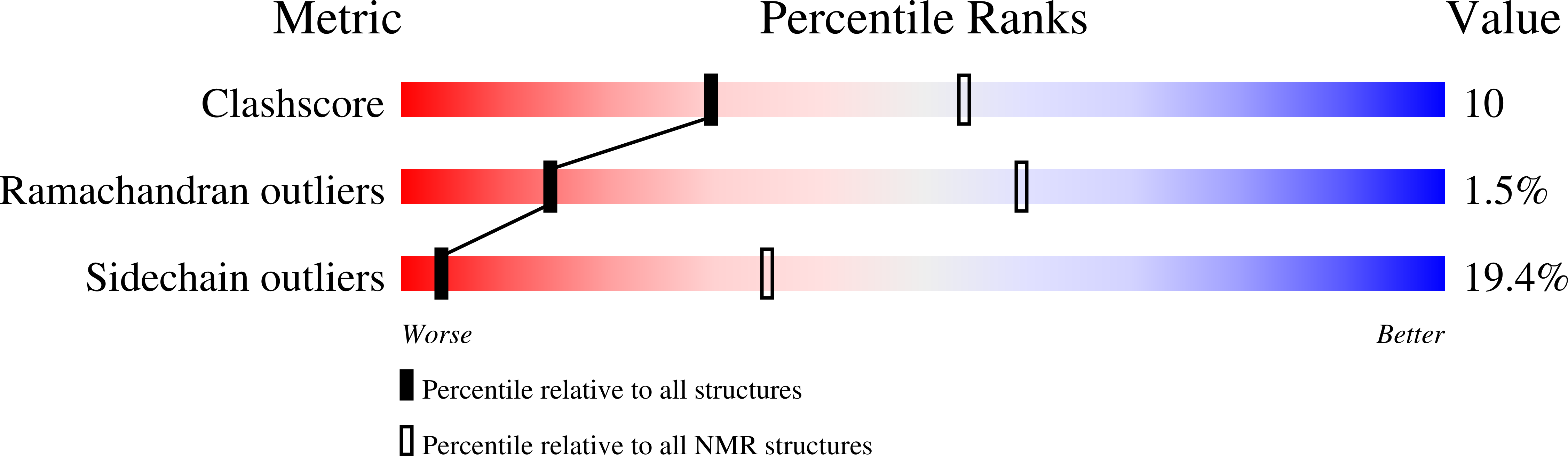
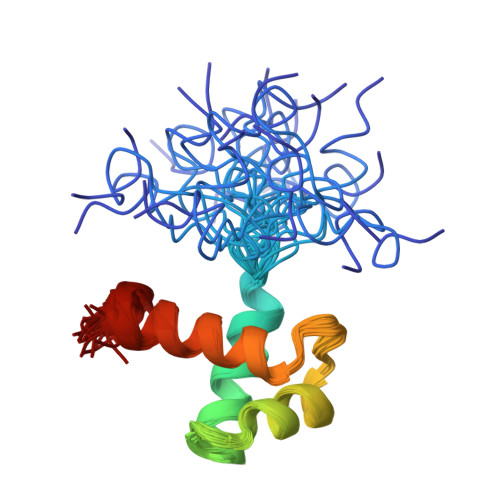
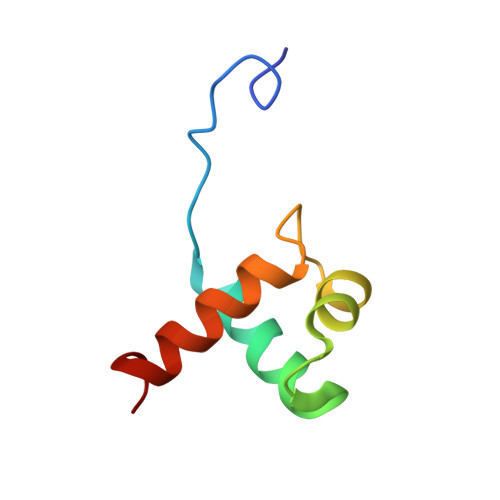
Comparison between TRF2 and TRF1 of their telomeric DNA-bound structures and DNA-binding activities
Hanaoka, S., Nagadoi, A., Nishimura, Y.(2005) Protein Sci 14: 119-130
- PubMed: 15608118
- DOI: https://doi.org/10.1110/ps.04983705
- Primary Citation of Related Structures:
1VF9, 1VFC - PubMed Abstract:
Mammalian telomeres consist of long tandem arrays of double-stranded telomeric TTAGGG repeats packaged by the telomeric DNA-binding proteins TRF1 and TRF2. Both contain a similar C-terminal Myb domain that mediates sequence-specific binding to telomeric DNA. In a DNA complex of TRF1, only the single Myb-like domain consisting of three helices can bind specifically to double-stranded telomeric DNA. TRF2 also binds to double-stranded telomeric DNA. Although the DNA binding mode of TRF2 is likely identical to that of TRF1, TRF2 plays an important role in the t-loop formation that protects the ends of telomeres. Here, to clarify the details of the double-stranded telomeric DNA-binding modes of TRF1 and TRF2, we determined the solution structure of the DNA-binding domain of human TRF2 bound to telomeric DNA; it consists of three helices, and like TRF1, the third helix recognizes TAGGG sequence in the major groove of DNA with the N-terminal arm locating in the minor groove. However, small but significant differences are observed; in contrast to the minor groove recognition of TRF1, in which an arginine residue recognizes the TT sequence, a lysine residue of TRF2 interacts with the TT part. We examined the telomeric DNA-binding activities of both DNA-binding domains of TRF1 and TRF2 and found that TRF1 binds more strongly than TRF2. Based on the structural differences of both domains, we created several mutants of the DNA-binding domain of TRF2 with stronger binding activities compared to the wild-type TRF2.
Organizational Affiliation:
Graduate School of Integrated Science, Yokohama City University, 1-7-29 Suehiro-cho, Tsurumi-ku, Yokohama, 230-0045, Japan.