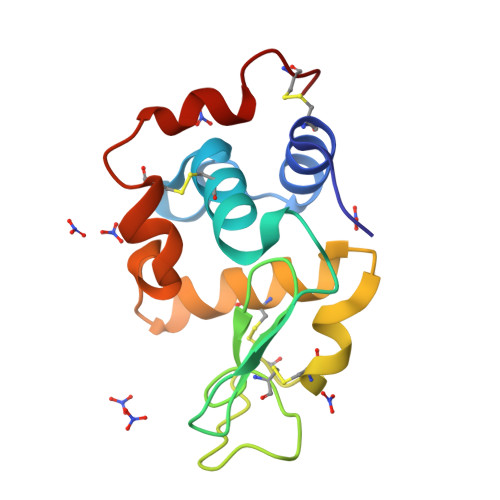
Phase transition of triclinic hen egg-white lysozyme crystal associated with sodium binding.
Harata, K., Akiba, T.(2004) Acta Crystallogr D Biol Crystallogr 60: 630-637
- PubMed: 15039550
- DOI: https://doi.org/10.1107/S0907444904001805
- Primary Citation of Related Structures:
1V7S, 1V7T - PubMed Abstract:
A triclinic crystal of hen egg-white lysozyme obtained from a D2O solution at 313 K was transformed into a new triclinic crystal by slow release of solvent under a temperature-regulated nitrogen-gas stream. The progress of the transition was monitored by X-ray diffraction. The transition started with the appearance of strong diffuse streaks. The diffraction spots gradually fused and faded with the emergence of diffraction from the new lattice; the scattering power of the crystal fell to a resolution of 1.5 A from the initial 0.9 A resolution. At the end of the transition, the diffuse streaks disappeared and the scattering power recovered to 1.1 A resolution. The transformed crystal contained two independent molecules and the solvent content had decreased to 18% from the 32% solvent content of the native crystal. The structure was determined at 1.1 A resolution and compared with the native structure refined at the same resolution. The backbone structures of the two molecules in the transformed crystal were superimposed on the native structure with root-mean-square deviations of 0.71 and 0.96 A. A prominent structural difference was observed in the loop region of residues Ser60-Leu75. In the native crystal, a water molecule located at the centre of this helical loop forms hydrogen bonds to main-chain peptide groups. In the transformed crystal, this water molecule is replaced by a sodium ion with octahedral coordination that involves water molecules and a nitrate ion. The peptide group connecting Arg73 and Asn74 is rotated by 180 degrees so that the CO group of Arg73 can coordinate to the sodium ion. The change in the X-ray diffraction pattern during the phase transition suggests that the transition proceeds at the microcrystal level. A mechanism is proposed for the crystal transformation.
Organizational Affiliation:
Biological Information Research Center, National Institute of Advanced Industrial Science and Technology, Central 6, 1-1-1 Higashi, Tsukuba, Ibaraki 305-8566, Japan. k-harata@aist.go.jp