Structure and Evolution of the Serum Paraoxonase Family of Detoxifying and Anti-Atherosclerotic Enzymes
Harel, M., Aharoni, A., Gaidukov, L., Brumshtein, B., Khersonsky, O., Meged, R., Dvir, H., Ravelli, R.B.G., Mccarthy, A., Toker, L., Silman, I., Sussman, J.L., Tawfik, D.S.(2004) Nat Struct Mol Biol 11: 412
- PubMed: 15098021
- DOI: https://doi.org/10.1038/nsmb767
- Primary Citation of Related Structures:
1V04 - PubMed Abstract:
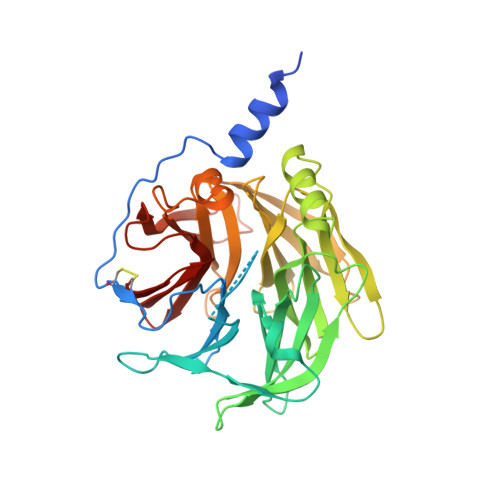
Members of the serum paraoxonase (PON) family have been identified in mammals and other vertebrates, and in invertebrates. PONs exhibit a wide range of physiologically important hydrolytic activities, including drug metabolism and detoxification of nerve agents. PON1 and PON3 reside on high-density lipoprotein (HDL, 'good cholesterol') and are involved in the prevention of atherosclerosis. We describe the first crystal structure of a PON family member, a variant of PON1 obtained by directed evolution, at a resolution of 2.2 A. PON1 is a six-bladed beta-propeller with a unique active site lid that is also involved in HDL binding. The three-dimensional structure and directed evolution studies permit a detailed description of PON1's active site and catalytic mechanism, which are reminiscent of secreted phospholipase A2, and of the routes by which PON family members diverged toward different substrate and reaction selectivities.
- Department of Structural Biology, The Weizmann Institute of Science, Rehovot 76 100, Israel.
Organizational Affiliation: