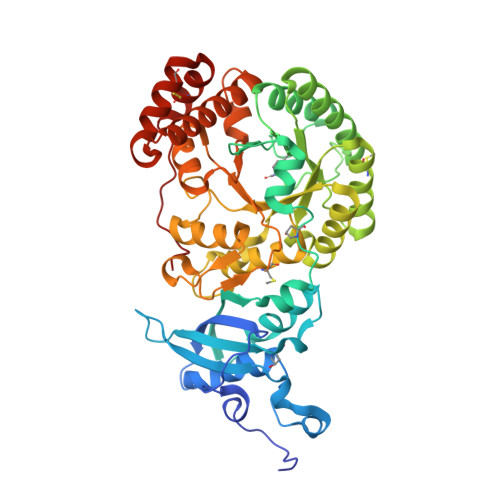
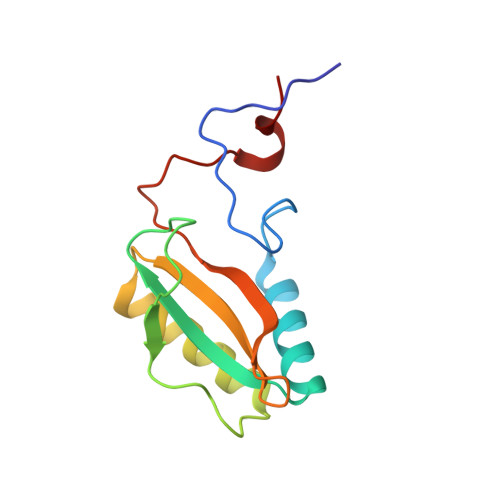
Chimeric Small Subunits Influence Catalysis without Causing Global Conformational Changes in the Crystal Structure of Ribulose-1,5-Bisphosphate Carboxylase/Oxygenase
Karkehabadi, S., Peddi, S.R., Anwaruzzaman, M., Taylor, T.C., Cederlund, A., Genkov, T., Andersson, I., Spreitzer, R.J.(2005) Biochemistry 44: 9851
- PubMed: 16026157
- DOI: https://doi.org/10.1021/bi050537v
- Primary Citation of Related Structures:
1UZD, 1UZH - PubMed Abstract:
Comparison of subunit sequences and X-ray crystal structures of ribulose-1,5-bisphosphate carboxylase/oxygenase indicates that the loop between beta-strands A and B of the small subunit is one of the most variable regions of the holoenzyme. In prokaryotes and nongreen algae, the loop contains 10 residues. In land plants and green algae, the loop is comprised of approximately 22 and 28 residues, respectively. Previous studies indicated that the longer betaA-betaB loop was required for the assembly of cyanobacterial small subunits with plant large subunits in isolated chloroplasts. In the present study, chimeric small subunits were constructed by replacing the loop of the green alga Chlamydomonas reinhardtii with the sequences of Synechococcus or spinach. When these engineered genes were transformed into a Chlamydomonas mutant that lacks small-subunit genes, photosynthesis-competent colonies were recovered, indicating that loop size is not essential for holoenzyme assembly. Whereas the Synechococcus loop causes decreases in carboxylation V(max), K(m)(O(2)), and CO(2)/O(2) specificity, the spinach loop causes complementary decreases in carboxylation V(max), K(m)(O(2)), and K(m)(CO(2)) without a change in specificity. X-ray crystal structures of the engineered proteins reveal remarkable similarity between the introduced betaA-betaB loops and the respective loops in the Synechococcus and spinach enzymes. The side chains of several large-subunit residues are altered in regions previously shown by directed mutagenesis to influence CO(2)/O(2) specificity. Differences in the catalytic properties of divergent Rubisco enzymes may arise from differences in the small-subunit betaA-betaB loop. This loop may be a worthwhile target for genetic engineering aimed at improving photosynthetic CO(2) fixation.
- Department of Molecular Biology, Swedish University of Agricultural Sciences, 751 24 Uppsala, Sweden.
Organizational Affiliation: