The Truncated Oxygen-Avid Hemoglobin from Bacillus Subtilis: X-Ray Structure and Ligand Binding Properties
Giangiacomo, L., Ilari, A., Boffi, A., Morea, V., Chiancone, E.(2005) J Biological Chem 280: 9192
- PubMed: 15590662
- DOI: https://doi.org/10.1074/jbc.M407267200
- Primary Citation of Related Structures:
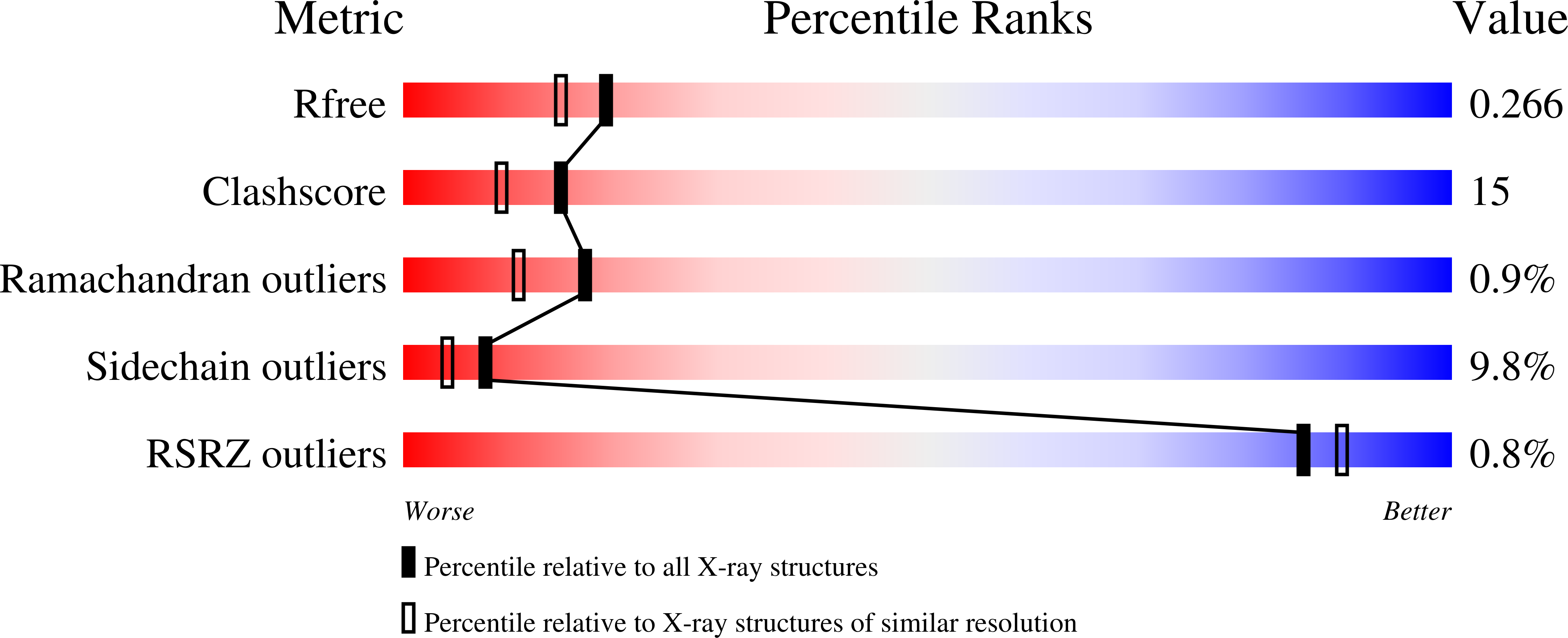
1UX8 - PubMed Abstract:
The group II truncated hemoglobin from Bacillus subtilis has been cloned, expressed, purified, and characterized. B. subtilis truncated hemoglobin is a monomeric protein endowed with an unusually high oxygen affinity (in the nanomolar range) such that the apparent thermodynamic binding constant for O2 exceeds that for CO by 1 order of magnitude. The kinetic basis of the high oxygen affinity resides mainly in the very slow rate of ligand release. The extremely stable ferrous oxygenated adduct is resistant to oxidation, which can be achieved only with oxidant in large excess, e.g. ferricyanide in 50-fold molar excess. The three-dimensional crystal structure of the cyano-Met derivative was determined at 2.15 A resolution. Although the overall fold resembles that of other truncated hemoglobins, the distal heme pocket displays a unique array of hydrophilic side chains in the topological positions that dominate the steric interaction with iron-bound ligands. In fact, the Tyr-B10, Thr-E7, and Gln-E11 oxygens on one side of the heme pocket and the Trp-G8 indole NE1 nitrogen on the other form a novel pattern of the "ligand-inclusive hydrogen bond network" described for mycobacterial HbO. On the proximal side, the histidine residue is in an unstrained conformation, and the iron-His bond is unusually short (1.91 A).
Organizational Affiliation:
Department of Biochemical Sciences and CNR Institute of Molecular Biology and Pathology, University of Rome La Sapienza, Piazzale Aldo Moro 5, 00185 Rome, Italy.