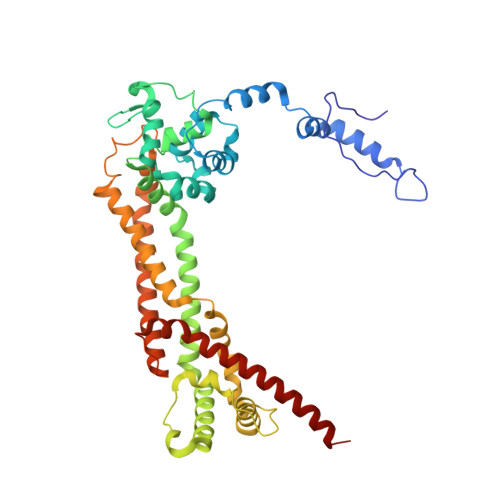
Crystal Structures of a Formin Homology-2 Domain Reveal a Tethered Dimer Architecture
Xu, Y., Moseley, J.B., Sagot, I., Poy, F., Pellman, D., Goode, B.L., Eck, M.J.(2004) Cell 116: 711
- PubMed: 15006353
- DOI: https://doi.org/10.1016/s0092-8674(04)00210-7
- Primary Citation of Related Structures:
1UX4, 1UX5 - PubMed Abstract:
Formin proteins participate in a wide range of cytoskeletal processes in all eukaryotes. The defining feature of formins is a highly conserved approximately 400 residue region, the Formin Homology-2 (FH2) domain, which has recently been found to nucleate actin filaments. Here we report crystal structures of the S. cerevesiae Bni1p FH2 domain. The mostly alpha-helical FH2 domain forms a unique "tethered dimer" in which two elongated actin binding heads are tied together at either end by an unusual lasso and linker structure. Biochemical and crystallographic observations indicate that the dimer is stable but flexible, with flexibility between the two halves of the dimer conferred by the linker segments. Although each half of the dimer is competent to interact with filament ends, the intact dimer is required for actin nucleation and processive capping. The tethered dimer architecture may allow formins to stair-step on the barbed end of an elongating nascent filament.
Organizational Affiliation:
Department of Cancer Biology, Dana-Farber Cancer Institute, Boston, MA 02115, USA.