Solution Structure and DNA Binding of the Effector Domain from the Global Regulator Prra(Rega) from Rhodobacter Sphaeroides: Insights Into DNA Binding Specificity
Laguri, C., Phillips-Jones, M.K., Williamson, M.P.(2003) Nucleic Acids Res 31: 6778
- PubMed: 14627811
- DOI: https://doi.org/10.1093/nar/gkg891
- Primary Citation of Related Structures:
1UMQ - PubMed Abstract:
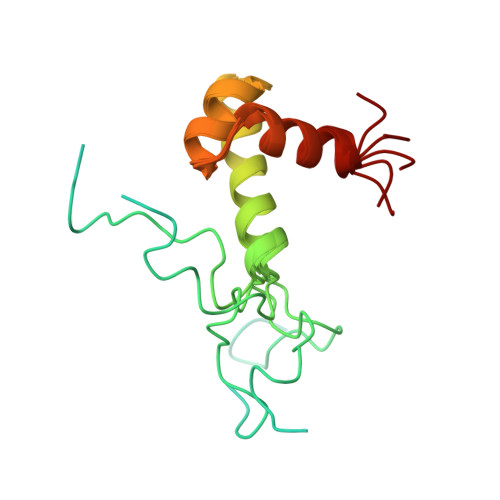
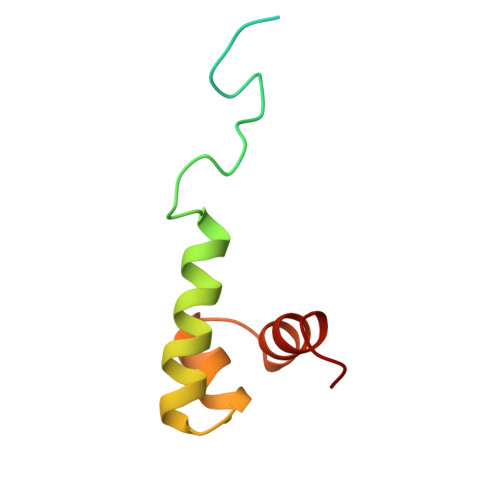
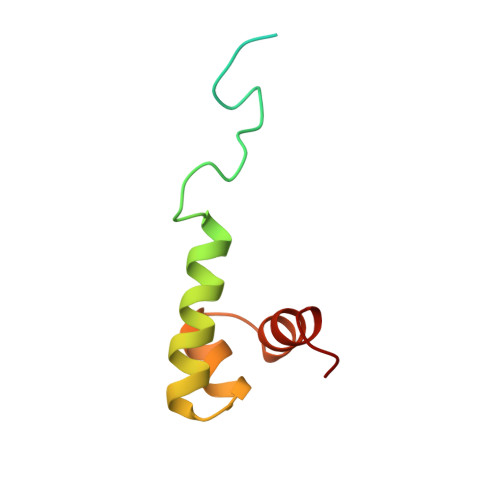
Prr/RegA response regulator is a global transcription regulator in purple bacteria Rhodobacter sphaeroides and Rhodobacter capsulatus, and is essential in controlling the metabolic changes between aerobic and anaerobic environments. We report here the structure determination by NMR of the C-terminal effector domain of PrrA, PrrAC. It forms a three-helix bundle containing a helix-turn-helix DNA binding motif. The fold is similar to FIS protein, but the domain architecture is different from previously characterised response regulator effector domains, as it is shorter than any characterised so far. Alignment of Prr/RegA DNA targets permitted a refinement of the consensus sequence, which contains two GCGNC inverted repeats with variable half-site spacings. NMR titrations of PrrAC with specific and non-specific DNA show which surfaces are involved in DNA binding and suggest residues important for binding specificity. A model of the PrrAC/DNA complex was constructed in which two PrrAC molecules are bound to DNA in a symmetrical manner.
Organizational Affiliation:
Department of Molecular Biology and Biotechnology, University of Sheffield, Firth Court, Western Bank, Sheffield S10 2UH, UK.