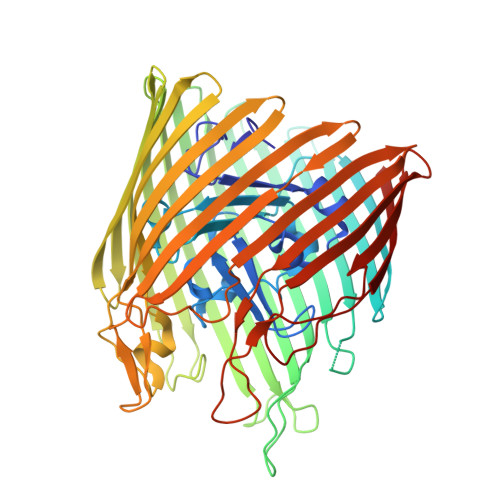
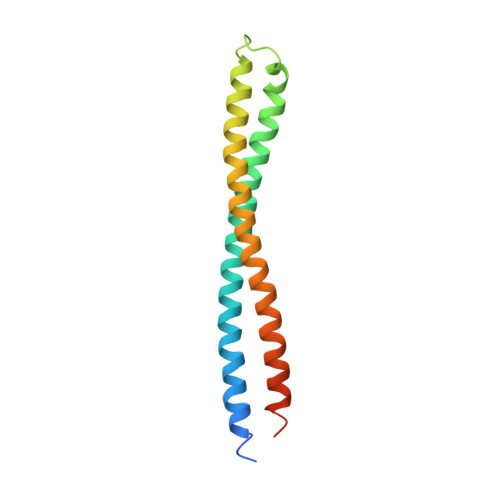
The structure of BtuB with bound colicin E3 R-domain implies a translocon
Kurisu, G., Zakharov, S.D., Zhalnina, M.V., Bano, S., Eroukova, V.Y., Rokitskaya, T.I., Antonenko, Y.N., Wiener, M.C., Cramer, W.A.(2003) Nat Struct Biol 10: 948-954
- PubMed: 14528295
- DOI: https://doi.org/10.1038/nsb997
- Primary Citation of Related Structures:
1UJW - PubMed Abstract:
Cellular import of colicin E3 is initiated by the Escherichia coli outer membrane cobalamin transporter, BtuB. The 135-residue 100-A coiled-coil receptor-binding domain (R135) of colicin E3 forms a 1:1 complex with BtuB whose structure at a resolution of 2.75 A is reported. Binding of R135 to the BtuB extracellular surface (DeltaG(o) = -12 kcal mol(-1)) is mediated by 27 residues of R135 near the coiled-coil apex. Formation of the R135-BtuB complex results in unfolding of R135 N- and C-terminal ends, inferred to be important for unfolding of the colicin T-domain. Small conformational changes occur in the BtuB cork and barrel domains but are insufficient to form a translocation channel. The absence of a channel and the peripheral binding of R135 imply that BtuB serves to bind the colicin, and that the coiled-coil delivers the colicin to a neighboring outer membrane protein for translocation, thus forming a colicin translocon. The translocator was concluded to be OmpF from the occlusion of OmpF channels by colicin E3.
Organizational Affiliation:
Department of Biological Sciences, Purdue University, Lilly Hall of Life Sciences, 915 W. State St., West Lafayette, Indiana 47907-1392, USA.