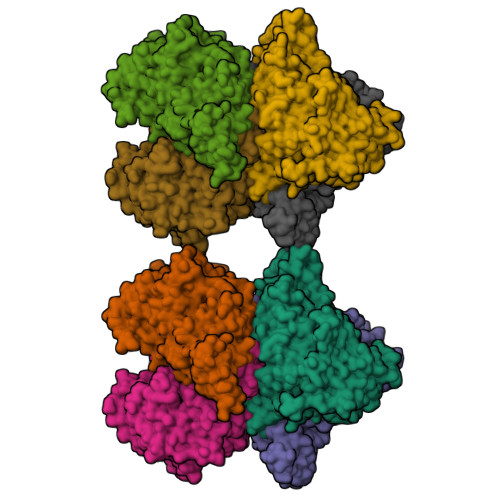
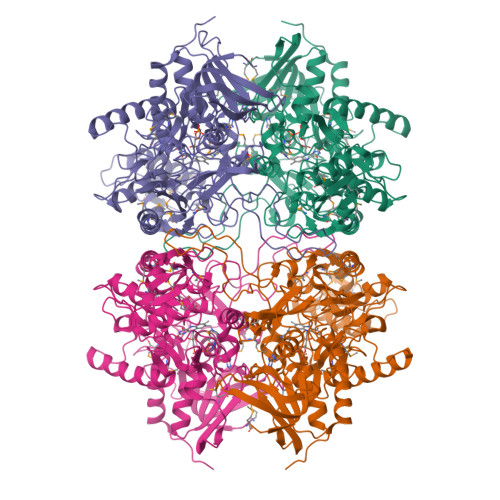
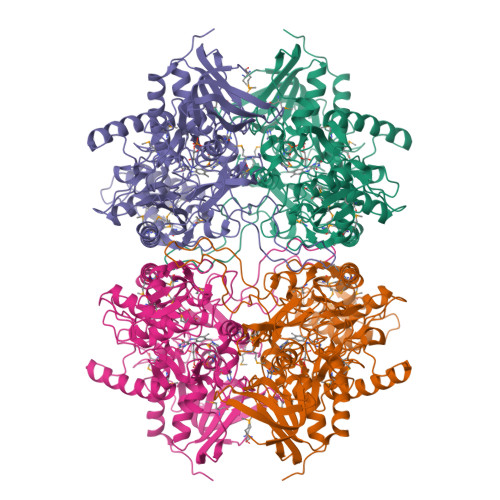
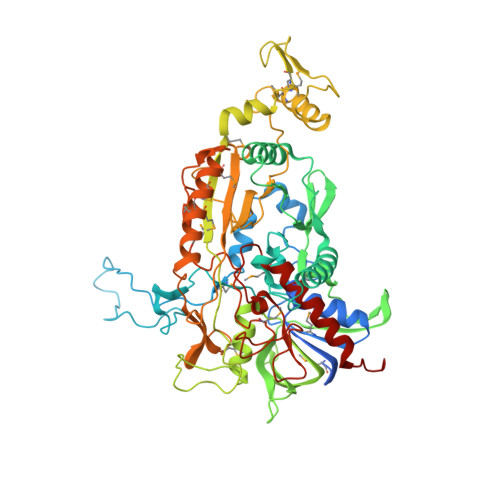
Crystal structure of pyranose 2-oxidase from the white-rot fungus peniophora sp.
Bannwarth, M., Bastian, S., Heckmann-Pohl, D., Giffhorn, F., Schulz, G.E.(2004) Biochemistry 43: 11683-11690
- PubMed: 15362852
- DOI: https://doi.org/10.1021/bi048609q
- Primary Citation of Related Structures:
1TZL - PubMed Abstract:
Pyranose 2-oxidase catalyzes the oxidation of a number of carbohydrates using dioxygen. The enzyme forms a D(2) symmetric homotetramer and contains one covalently bound FAD per subunit. The structure of the enzyme from Peniophora sp. was determined by multiwavelength anomalous diffraction (MAD) based on 96 selenium sites per crystallographic asymmetric unit and subsequently refined to good-quality indices. According to its chain fold, the enzyme belongs to the large glutathione reductase family and, in a more narrow sense, to the glucose-methanol-choline oxidoreductase (GMC) family. The tetramer contains a spacious central cavity from which the substrate enters one of the four active centers by penetrating a mobile barrier. Since this cavity can only be accessed by glucose-sized molecules, the enzyme does not convert sugars that are part of a larger molecule. The geometry of the active center and a comparison with an inhibitor complex of the homologous enzyme cellobiose dehydrogenase allow the modeling of the reaction at a high confidence level.
Organizational Affiliation:
Institut für Organische Chemie und Biochemie, Albert-Ludwigs-Universität, Albertstrasse 21, 79104 Freiburg im Breisgau, Germany.