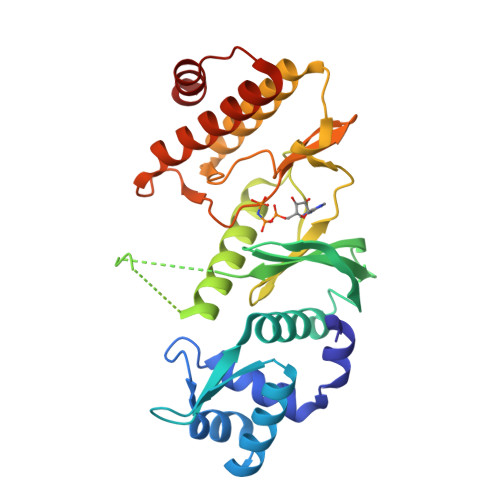
Crystal Structure of A. fulgidus Rio2 Defines a New Family of Serine Protein Kinases
LaRonde-LeBlanc, N., Wlodawer, A.(2004) Structure 12: 1585-1594
- PubMed: 15341724
- DOI: https://doi.org/10.1016/j.str.2004.06.016
- Primary Citation of Related Structures:
1TQI, 1TQM, 1TQP - PubMed Abstract:
The RIO family of atypical serine/threonine kinases contains two subfamilies, Rio1 and Rio2, highly conserved from archaea to man. Both RIO proteins from Saccharomyces cerevisiae catalyze serine phosphorylation in vitro, and the presence of conserved catalytic residues is required for cell viability. The activity of Rio2 is necessary for rRNA cleavage in 40S ribosomal subunit maturation. We solved the X-ray crystal structure of Archaeoglobus fulgidus Rio2, with and without bound nucleotides, at 2.0 A resolution. The C-terminal RIO domain is indeed structurally homologous to protein kinases, although it differs from known serine kinases in ATP binding and lacks the regions important for substrate binding. Unexpectedly, the N-terminal Rio2-specific domain contains a winged helix fold, seen primarily in DNA-binding proteins. These discoveries have implications in determining the target and function of RIO proteins and define a distinct new family of protein kinases.
Organizational Affiliation:
Protein Structure Section, Macromolecular Crystallography Laboratory, National Cancer Institute, NCI-Frederick, MD 21702, USA.